Abstract
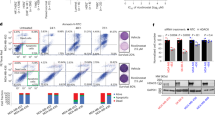
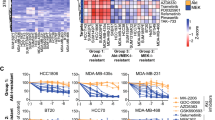
We leverage genomic and biochemical data to identify synergistic drug regimens for breast cancer. In order to study the mechanism of the histone deacetylase (HDAC) inhibitors valproic acid (VPA) and suberoylanilide hydroxamic acid (SAHA) in breast cancer, we generated and validated genomic profiles of drug response using a series of breast cancer cell lines sensitive to each drug. These genomic profiles were then used to model drug response in human breast tumors and show significant correlation between VPA and SAHA response profiles in multiple breast tumor data sets, highlighting their similar mechanism of action. The genes deregulated by VPA and SAHA converge on the cell cycle pathway (Bayes factor 5.21 and 5.94, respectively; P-value 10−8.6 and 10−9, respectively). In particular, VPA and SAHA upregulate key cyclin-dependent kinase (CDK) inhibitors. In two independent datasets, cancer cells treated with CDK inhibitors have similar gene expression profile changes to the cellular response to HDAC inhibitors. Together, these results led us to hypothesize that VPA and SAHA may interact synergistically with CDK inhibitors such as PD-033299. Experiments show that HDAC and CDK inhibitors have statistically significant synergy in both breast cancer cell lines and primary 3-dimensional cultures of cells from pleural effusions of patients. Therefore, synergistic relationships between HDAC and CDK inhibitors may provide an effective combinatorial regimen for breast cancer. Importantly, these studies provide an example of how genomic analysis of drug–response profiles can be used to design rational drug combinations for cancer treatment.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 6 print issues and online access
$259.00 per year
only $43.17 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Altekruse S, Kosary C, Krapcho M, Neyman N, Aminou R, Waldron W et al. SEER Cancer Statistics Review 1975–2007. National Cancer Institute: Bethesda, MD. http://seer.cancer.gov/csr/1975_2007/, Access based on November 2009 SEER data submission, posted to the SEER website, 2010.
Woodcock J, Griffin JP, Behrman RE . Development of novel combination therapies. N Engl J Med 2011; 364: 985–987.
Hanahan D, Weinberg RA . The hallmarks of cancer. Cell 2000; 100: 57–70.
Lund AH, van Lohuizen M . Epigenetics and cancer. Genes Dev 2004; 18: 2315–2335.
Baylin SB, Ohm JE . Epigenetic gene silencing in cancer—a mechanism for early oncogenic pathway addiction? Nat Rev Cancer 2006; 6: 107–116.
Roth SY, Denu JM, Allis CD . Histone acetyltransferases. Annu Rev Biochem 2001; 70: 81–120.
Marks PA, Jiang X . Histone deacetylase inhibitors in programmed cell death and cancer therapy. Cell Cycle 2005; 4: 549–551.
Bolden JE, Peart MJ, Johnstone RW . Anticancer activities of histone deacetylase inhibitors. Nat Rev Drug Discov 2006; 5: 769–784.
Zhang Z, Yamashita H, Toyama T, Sugiura H, Omoto Y, Ando Y et al. HDAC6 expression is correlated with better survival in breast cancer. Clin Cancer Res 2004; 10: 6962–6968.
Wilson AJ, Byun DS, Popova N, Murray LB, L′Italien K, Sowa Y et al. Histone deacetylase 3 (HDAC3) and other class I HDACs regulate colon cell maturation and p21 expression and are deregulated in human colon cancer. J Biol Chem 2006; 281: 13548–13558.
Zhang Z, Yamashita H, Toyama T, Sugiura H, Ando Y, Mita K et al. Quantitation of HDAC1 mRNA expression in invasive carcinoma of the breast*. Breast Cancer Res Treat 2005; 94: 11–16.
Halkidou K, Gaughan L, Cook S, Leung HY, Neal DE, Robson CN . Upregulation and nuclear recruitment of HDAC1 in hormone refractory prostate cancer. Prostate 2004; 59: 177–189.
Carew JS, Giles FJ, Nawrocki ST . Histone deacetylase inhibitors: mechanisms of cell death and promise in combination cancer therapy. Cancer Lett 2008; 269: 7–17.
Marks PA, Richon VM, Kelly WK, Chiao JH, Miller T . Histone deacetylase inhibitors: development as cancer therapy. Novartis Foundation Symposium 2004; 259: 269–281;, discussion 281-268.
Kelly WK, O′Connor OA, Marks PA . Histone deacetylase inhibitors: from target to clinical trials. Expert Opin Investig Drugs 2002; 11: 1695–1713.
Takai N, Desmond JC, Kumagai T, Gui D, Said JW, Whittaker S et al. Histone deacetylase inhibitors have a profound antigrowth activity in endometrial cancer cells. Clin Cancer Res 2004; 10: 1141–1149.
Haggarty SJ, Koeller KM, Wong JC, Grozinger CM, Schreiber SL . Domain-selective small-molecule inhibitor of histone deacetylase 6 (HDAC6)-mediated tubulin deacetylation. Proc Natl Acad Sci USA 2003; 100: 4389–4394.
Cucciolla V, Borriello A, Criscuolo M, Sinisi AA, Bencivenga D, Tramontano A et al. Histone deacetylase inhibitors upregulate p57Kip2 level by enhancing its expression through Sp1 transcription factor. Carcinogenesis 2008; 29: 560–567.
Faiola F, Liu X, Lo S, Pan S, Zhang K, Lymar E et al. Dual regulation of c-Myc by p300 via acetylation-dependent control of Myc protein turnover and coactivation of Myc-induced transcription. Mol Cell Biol 2005; 25: 10220–10234.
Monks A, Hose CD, Pezzoli P, Kondapaka S, Vansant G, Petersen KD et al. Gene expression-signature of belinostat in cell lines is specific for histone deacetylase inhibitor treatment, with a corresponding signature in xenografts. Anticancer Drugs 2009; 20: 682–692.
Glaser KB, Staver MJ, Waring JF, Stender J, Ulrich RG, Davidsen SK . Gene expression profiling of multiple histone deacetylase (HDAC) inhibitors: defining a common gene set produced by HDAC inhibition in T24 and MDA carcinoma cell lines. Mol Cancer Ther 2003; 2: 151–163.
Yang X, Karuturi RK, Sun F, Aau M, Yu K, Shao R et al. CDKN1C (p57) is a direct target of EZH2 and suppressed by multiple epigenetic mechanisms in breast cancer cells. PLoS One 2009; 4: e5011.
Kikuchi T, Toyota M, Itoh F, Suzuki H, Obata T, Yamamoto H et al. Inactivation of p57KIP2 by regional promoter hypermethylation and histone deacetylation in human tumors. Oncogene 2002; 21: 2741–2749.
Pateras IS, Apostolopoulou K, Koutsami M, Evangelou K, Tsantoulis P, Liloglou T et al. Downregulation of the KIP family members p27(KIP1) and p57(KIP2) by SKP2 and the role of methylation in p57(KIP2) inactivation in nonsmall cell lung cancer. Int J Cancer 2006; 119: 2546–2556.
Hayslip J, Montero A . Tumor suppressor gene methylation in follicular lymphoma: a comprehensive review. Mol Cancer 2006; 5: 44.
Algar EM, Muscat A, Dagar V, Rickert C, Chow CW, Biegel JA et al. Imprinted CDKN1C is a tumor suppressor in rhabdoid tumor and activated by restoration of SMARCB1 and histone deacetylase inhibitors. PLoS One 2009; 4: e4482.
Larson PS, Schlechter BL, King CL, Yang Q, Glass CN, Mack C et al. CDKN1C/p57kip2 is a candidate tumor suppressor gene in human breast cancer. BMC Cancer 2008; 8: 68.
Tallarida RJ . Drug synergism: its detection and applications. J Pharmacol Exp Ther 2001; 298: 865–872.
Iorio F, Bosotti R, Scacheri E, Belcastro V, Mithbaokar P, Ferriero R et al. Discovery of drug mode of action and drug repositioning from transcriptional responses. Proc Natl Acad Sci USA 2010; 107: 14621–14626.
Berkofsky-Fessler W, Nguyen TQ, Delmar P, Molnos J, Kanwal C, DePinto W et al. Preclinical biomarkers for a cyclin-dependent kinase inhibitor translate to candidate pharmacodynamic biomarkers in phase I patients. Mol Cancer Ther 2009; 8: 2517–2525.
Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science 2006; 313: 1929–1935.
Welsbie DS, Xu J, Chen Y, Borsu L, Scher HI, Rosen N et al. Histone deacetylases are required for androgen receptor function in hormone-sensitive and castrate-resistant prostate cancer. Cancer Res 2009; 69: 958–966.
Wood JR, Nelson-Degrave VL, Jansen E, McAllister JM, Mosselman S, Strauss III JF . Valproate-induced alterations in human theca cell gene expression: clues to the association between valproate use and metabolic side effects. Physiol Genomics 2005; 20: 233–243.
Meijer L, Borgne A, Mulner O, Chong JP, Blow JJ, Inagaki N et al. Biochemical and cellular effects of roscovitine, a potent and selective inhibitor of the cyclin-dependent kinases cdc2, cdk2 and cdk5. Eur J Biochem 1997; 243: 527–536.
Fry DW, Harvey PJ, Keller PR, Elliott WL, Meade M, Trachet E et al. Specific inhibition of cyclin-dependent kinase 4/6 by PD 0332991 and associated antitumor activity in human tumor xenografts. Mol Cancer Ther 2004; 3: 1427–1438.
Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell 2006; 10: 515–527.
Bissell MJ, Labarge MA . Context, tissue plasticity, and cancer: are tumor stem cells also regulated by the microenvironment? Cancer Cell 2005; 7: 17–23.
Griffith LG, Swartz MA . Capturing complex 3D tissue physiology in vitro. Nat Rev Mol Cell Biol 2006; 7: 211–224.
American Cancer Society. Cancer Facts & Figures. American Cancer Society: Atlanta, 2011.
Buzdar AU . Role of biologic therapy and chemotherapy in hormone receptor- and HER2-positive breast cancer. Ann Oncol 2009; 20: 993–999.
Munster P, Marchion D, Bicaku E, Lacevic M, Kim J, Centeno B et al. Clinical and biological effects of valproic acid as a histone deacetylase inhibitor on tumor and surrogate tissues: phase I/II trial of valproic acid and epirubicin/FEC. Clin Cancer Res 2009; 15: 2488–2496.
Travaglini L, Vian L, Billi M, Grignani F, Nervi C . Epigenetic reprogramming of breast cancer cells by valproic acid occurs regardless of estrogen receptor status. Int J Biochem Cell Biol 2009; 41: 225–234.
Chavez-Blanco A, Perez-Plasencia C, Perez-Cardenas E, Carrasco-Legleu C, Rangel-Lopez E, Segura-Pacheco B et al. Antineoplastic effects of the DNA methylation inhibitor hydralazine and the histone deacetylase inhibitor valproic acid in cancer cell lines. Cancer Cell Int 2006; 6: 2.
Candelaria M, Gallardo-Rincon D, Arce C, Cetina L, Aguilar-Ponce JL, Arrieta O et al. A phase II study of epigenetic therapy with hydralazine and magnesium valproate to overcome chemotherapy resistance in refractory solid tumors. Ann Oncol 2007; 18: 1529–1538.
Munster P, Marchion D, Bicaku E, Schmitt M, Lee JH, DeConti R et al. Phase I trial of histone deacetylase inhibition by valproic acid followed by the topoisomerase II inhibitor epirubicin in advanced solid tumors: a clinical and translational study. J Clin Oncol 2007; 25: 1979–1985.
Arce C, Perez-Plasencia C, Gonzalez-Fierro A, de la Cruz-Hernandez E, Revilla-Vazquez A, Chavez-Blanco A et al. A proof-of-principle study of epigenetic therapy added to neoadjuvant doxorubicin cyclophosphamide for locally advanced breast cancer. PLoS One 2006; 1: e98.
Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA et al. Molecular portraits of human breast tumours. Nature 2000; 406: 747–752.
Acknowledgements
This study was funded by the National Institute of Health (R01GM085601, AHB); the Pharmaceutical Research and Manufacturers of America (AHB); a Multidisciplinary Cancer Research Training Program award (T32 CA93247, RS and AC) and an award from the MIDT cancer center support grant. The Breast Interdisciplinary Group of the University of Utah is acknowledged for their assistance in breast tumor collection.
Author Contributions
AHB and RS designed the study. AHB, ALC and RS wrote the manuscript. RS and LC performed the in vitro experiments. ALC and YS performed the microarray analysis, profile generation and profile validation. ALC and AHB performed the gene ontology analysis. ALC, PM and AHB performed the data analysis, statistical analysis and synergy calculations.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the The Pharmacogenomics Journal website
Rights and permissions
About this article
Cite this article
Soldi, R., Cohen, A., Cheng, L. et al. A genomic approach to predict synergistic combinations for breast cancer treatment. Pharmacogenomics J 13, 94–104 (2013). https://doi.org/10.1038/tpj.2011.48
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/tpj.2011.48
Keywords
This article is cited by
-
Valproic acid inhibits cell growth in both MCF-7 and MDA-MB231 cells by triggering different responses in a cell type-specific manner
Journal of Translational Medicine (2023)
-
State-of-the-Art Technologies to Interrogate Genetic/Genomic Components of Drug Response
Current Genetic Medicine Reports (2013)