Abstract
“Fairy rings” resulting from fungus-stimulated plant growth occur all over the world. In 2010, 2-azahypoxanthine (AHX) from a fungus Lepista sordida was identified as the “fairy” that stimulates plant growth. Furthermore, 2-aza-8-oxohypoxanthine (AOH) was isolated as a common metabolite of AHX in plants, and the endogenous existence of AHX and AOH in plants was proved. The structure of AHX allowed us to hypothesize that AHX was derived from 5-aminoimidazole-4-carboxamide ribonucleotide (AICAR). Thus, we performed a feeding experiment that supplied AICAR to L. sordida. Consumption of AICAR and accumulation of AHX were observed after feeding. The mycelia extract had enzymatic activity of adenine/5-aminoimidazole-4-carboxamide phosphoribosyltransferase (APRT). APRT gene of L. sordida revealed its structural characteristics in homology modeling and showed transcriptional enhancement after feeding. These results support that AHX was synthesized from AICAR and AHX biosynthesis was transcriptionally controlled by AICAR, indicating the presence of novel purine metabolic pathway in L. sordida.
Similar content being viewed by others
Introduction
“Fairy rings” is a disease symptom in lawn; rapidly growing, lush green circular bands of grass and/or circles of mushrooms are observed1. It has been found that more than 60 of basidiomycete fungi form fairy rings1,2. Fairy ring disease has been found on all grass types all over the world3, particularly on golf courses and athletic fields. It is often observed in hot, dry and drought weather conditions. The disease symptoms shows plant withering, wilting in rings, or activation of plant growth such as dark green turf grass4, leading to stimulated, or no influence on growth5.
Lepista sordida (division: Basidiomycota, order: Agaricales) has been a targeted fungus to study the fairy ring formation mechanism in research. Fairy rings caused by L. sordida have been reported on Zoysia grass (Zoysia matrella and Zoysia japonica Steud)5. Recently, insight into the molecular mechanism of fairy ring formation lead to the isolation of a plant growth regulator, 2-azahypoxanthine (AHX), from the fungus6. AHX stimulated the growth of various kinds of plants including turfgrass6. Subsequently, 2-aza-8-oxohypoxanthine (AOH) was isolated from rice as a derivative metabolite of AHX7. Both the compounds increased grain yields of wheat and rice in field experiments8. AHX is chemically synthesized from 5-aminoimidazole-4-carboxamide (AICA); AICA reacts with NaNO2 and then with NH3, giving AHX7. AHX and its ribotide (AICAR) are common members on the purine metabolic pathway in animals, plants and microorganisms, and AICAR is a precursor of fundamental metabolites such as inosine monophosphate (IMP), inosine, hypoxanthine, xanthine and uric acid in the pathway. From our findings and the facts mentioned above, we hypothesized that plants themselves produce AHX through a pathway similar to the chemical synthesis, and we proved the hypothesis experimentally7. However, it remains unknown whether the pathway exists in fairy-ring forming fungi and how AICA is synthesized in cells.
Adenine/5-aminoimidazole-4-carboxamide phosphoribosyltransferase (APRT; EC 2.4.2.7) also called as nucleotide pyrophosphorylase is involved in AHX biosynthesis. APRT catalyzes the reversible transfer of the 5′-phosphoribosyl moiety from α-D-5-phosphoribosyl-1-pyrophosphate (PRPP) to adenine to form adenosine-5-monophosphate (AMP) and pyrophosphate (PPi)9. It also recognizes hypoxanthine, 2,6-diaminopurine, and AICA as substrates, although its binding affinities to the substrates are weaker compared to adenine10. Nucleotide pyrophosphorylase purified from beef liver converted AICAR and pyrophosphate to AICA and 5-phosphoribosyl pyrophosphate11. APRT is conserved in various organisms: animals, bacteria, and plant species12,13,14,15,16,17. The primary role of APRT is in adenine salvage and recycling18,19. In APRT-deficient human cells, adenine is converted to 2,8-dihydroxyadenine (DHA) by xanthine dehydrogenase, and accumulation of DHA is toxic to renal tubular and interstitial cells20. In the flowering plant, Arabidopsis, mutations in APRT affected pollen development causing male sterile21. APRT protein and its crystal structure from animals, bacteria, protista, fungi and archaea have been determined until now has been well-studied9,22,23,24,25,26,27. By contrast, there is still no knowledge on APRT in fairy ring-forming fungi.
The goal of this study is to examine whether AICAR could be a substrate for AHX biosynthesis in L. sordida. In this study, we provide answers for the three following points: 1) how feeding of AICAR in L. sordida mycelia culture affects on AHX accumulation and APRT gene expression, 2) what are the structural features of APRT, and 3) whether L. sordida mycelia has an enzyme activity for interconversion between AICAR and AICA or not.
Here, we performed a feeding experiment that supplied AICAR into L. sordida, determination of AICAR-converting activity of the crude enzyme extract from the mycelia, identification and isolation of an APRT gene in the fungus, and investigation of expression levels of the APRT gene during the feeding experiment.
Results
Feeding of AICAR into mycelia culture
In the hypothesized AHX/AOH biosynthetic pathway, AICAR is the initial precursor through AICA (Fig. 1). If this hypothesis is true, provision of AICAR to L. sordida should activate accumulation of AHX in the fungus. Therefore, we fed AICAR to L. sordida culture and quantified the amounts of AICAR and AHX until 48 h after feeding. AICAR concentration of non-fed control was between 81.5 ± 7.8 and 111.0 ± 11.5 μM during the experiment. The feeding increased the concentration of AICAR to 232.8 ± 9.8 μM from 87.8 ± 12.1 μM at 0 h, and then continuously decreased until 48 h after feeding (Fig. 2A). In comparison of these two samples, significant difference was observed at 12 h after feeding (Fig. 2A). These results indicated that a large part of AICAR fed was consumed within 24 h after feeding. However, no significant difference was observed in the non-fed control of AHX, its concentration decreased to 375.8 ± 30.8 μM from 454.5 ± 32.8 μM at 12 h after feeding. The feeding of AICAR slightly increased AHX accumulation to 475.9 ± 35.2 μM at 12 h after feeding, and decreased to 401.3 ± 28.1 μM at 24 h after feeding. Comparing these two results show significant difference between fed and non-fed groups at 12 h after feeding (P ≤ 0.05 at 12 h). All the results indicated that there exists a correlation between consumption of AICAR and accumulation of AHX in the mycelia culture at 12 h after feeding and AICAR was utilized for AHX biosynthesis in the mycelia.
Transitional changes of AICAR and AHX contents in mycelia culture after feeding of AICAR.
Error bar represents standard deviation of five biological replicates. Asterisk represents statistical significance of difference from control at 5% level. (A) AICAR. (B) AHX. The sample after feeding of AICAR is indicated with white circles and control (without AICAR) is indicated with black circles.
Enzymatic activity assay in mycelia
Crude extract from the mycelia culture was prepared and tested for AICAR conversion to AICA in the extract, using LC-MS/MS. As shown in Fig. 3, the crude extract with AICAR produced AICA. Whereas, AICA was not detected in reactions without either substrate or enzyme (Fig. 3B and D). These results showed that mycelia contained enzyme(s) converts AICAR to AICA.
Analysis of APRT activity in crude enzyme extract by LC-MS/MS.
Multiple reaction monitoring (MRM) for AICA including MS (left side) and MS/MS spectra (right side). (A) AICA standard. (B) Buffer (100 mM Tris-HCl, pH 8.3) and AICAR. (C) Crude enzyme extract and AICAR. (D) Crude enzyme extract and distilled water. Arrow indicates a signal of AICA.
Identification of APRT gene
To explore APRT genes in L. sordida, genomic scaffolds were generated by next-generation sequencing approach. A hybrid sequencing strategy by 454 pyrosequencing and Illumina sequencing techniques was employed. For 454 pyrosequencing, 915,457 of reads were generated. For Illumina sequencing, 66,506,146 and 71,023,220 short reads were generated from the PE-library and MP-library, respectively. The short reads were preprocessed to extract 79,179,992 of high-quality reads in total (See details in Materials and Methods and Supplementary Table 1). The data were conducted to a hybrid assembly strategy (See details in Materials and Methods). As a result, 812 of genomic scaffolds were constructed (Supplementary Table 2). The scaffold sequences were then applied to a search for APRT gene. Namely, we queried an APRT sequence in yeast (Genbank ID: L14434.1) by nhmmer search against the genomic scaffolds. The search identified a genomic scaffold including a potential APRT gene in L. sordida as a top hit with statistical significance (E-value of 8E-4) (Supplementary Fig. 1A). The second hit showed no statistical significance (E-value of 5.3). By aligning the APRT gene sequence with those of Coprinopsis cinerea and Moniliophthora roreri (Genbank ID: XM_001837049 and XM_007844891, respectively), the coding region was predicted (Supplementary Fig. 1B).
cDNA cloning of APRT in L. sordida
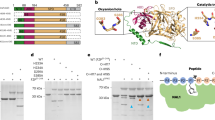
cDNA of APRT was isolated from the mycelia of L. sordida. The cDNA amplified by RT-PCR was cloned into a vector pMD20-T sequenced (deposited to DDBJ, accession number LC060066). cDNA sequence perfectly matched with the predicted coding sequence from the genomic scaffold (data not shown). The open reading frame was 546 bp length, and it coded 182 amino acid protein (Supplementary Fig. 2). The deduced amino acid sequence showed significant homologies with those of APRTs in Saccharomyces cerevisiae (51%), Drosophila pseudoobscura (43%), Mus musculus (44%), Rattus norvegicus (41%), Homo sapiens (40%), Arabidopsis thaliana (44% and 38%), Escherichia coli (40%), and Leishmania donovani (32%).9,14,15,16,23,28,29,30,31. We observed core motif sequences of APRT in the L. sordida APRT (Fig. 4). Phosphoribosyl pyrophosphate (PRPP)-binding motifs, composed of citrate contact region in crystal structures of L. donovani APRT23 and 5′-phosphate contact region in S. cerevisiae APRT9, were completely conserved in L. sordida. The amino acid position of 129 and 137, residues of Mg2+ binding were also conserved in APRTs31,32,33,34,35,36.
A multiple alignment of APRTs in 13 species.
Conserved sequence in all the APRT and 12 APRT are colored in yellow and blue, respectively. Residues involved in 5′-phosphate binding and Mg-pyrophosphate binding were circled in red and in black, respectively. The following APRTs were applied (GenBank accession numbers in parentheses): P.involutu, Paxillus involutus (KIJ20178.1); M.roreri, Moniliophthora roreri (XM_007844891.1); C. cinerea, Coprinopsis cinerea (XM_001837049.1); S. cerevi, Saccharomyces cerevisiae (P49435); D. pseudo, Drosophila pseudoobscura (L06281.1); M. musculus, Mus musculus (NM_009698.2); R. norvegi, Rattus norvegicus (NM_001013061.1); H. sapiens, Homo sapiens (P07741); A. thali-1, Arabidopsis thaliana (L19637.1); A. thali-2, Arabidopsis thaliana (X96867.1); E. coli, Escherichia coli (M14040.1); L. Donovani, Leishmania Donovani; (1QB7_A).
In structural analysis of the APRT, predicted five of α-helix and nine of β-strand structures seen in S. cerevisiae were also observed in the APRT of L. sordida (Fig. 4). A missing region (51-Thr-Ile-Thr-Lys-54), which was conserved among fungi, was also present. This region formed a part of loop in homology modeling with APRT in S. cerevisiae by SWISS-MODEL (Supplementary Fig. 3). The insertion of the missing region in APRT in L. sordida provided a larger space inside of the loop. Notably, the missing region contained a distinctive conserved amino acid Lys-54 between L. sordida and a fairy ring-forming fungi Paxillus involutus (Fig. 4). ProtScale analysis represented the loop region was more hydrophilic than that of S. cerevisiae (Supplementary Fig. 4). These results suggested that the loop in L. sordida has high flexibility.
Gene expression analysis of APRT and AICARFT gene
We examined the effect of AICAR feeding on APRT and AICARFT at mRNA level. Gene expression of APRT and AICARFT in mycelia culture was analyzed by qRT-PCR. In gene expression of APRT, the relative expression level of control did not change significantly. In contrast, APRT started to increase at 12 h after feeding and maintained at high levels until 36 h after feeding (Fig. 5A). The response was consistent with AHX accumulation at 12 h after feeding (Fig. 2B). The AICARFT expression level of non-fed control was between 0.84 ± 0.05 and 1.4 ± 0.3 during the experiment. Although there was no significant difference of AICARFT expression level between fed and non-fed groups, expression level of AICAR fed groups tended to slightly decrease at 12 h after feeding. These results suggested that AICARFT constantly expressed and the significant response to AICAR feeding was not observed in L. sordida mycelia culture.
Gene expression of APRT and AICARFT in response to AICAR feeding.
X-axis represents normalized gene expression level of the APRT (A) and AICARFT (B) by an actin. Error bar indicates standard deviation of four biological replicates. Asterisk represents significant difference to from negative control (P < 0.05 by Student’s T-test). The sample after feeding of AICAR is indicated with white circles and control (without AICAR) is indicated with black circles.
Discussion
In this study, we established a feeding experiment procedure in L. sordida mycelia culture for exploring enzyme genes in AHX and AOH biosynthetic pathway. After feeding of AICAR to the mycelia culture, consumption of AICAR and accumulation of AHX were observed in the culture (Fig. 2). In the feeding experiment, the APRT gene enhanced after 12 h of feeding (Fig. 5). These results suggested that the APRT gene acted upon AHX and AOH biosynthesis. Moreover, the presence of AICAR-converting enzyme in the crude enzyme extract of L. sordida mycelia was demonstrated by detecting AICA in LC-MS analysis. Our data suggested that the APRT gene responded to addition of AICAR and also might be involved in converting AICAR to AICA in the mycelia extract.
Despite the drastic reduction of AICAR until 48 h after feeding, clear increase of AHX was only observed at 12 h after feeding. But, gene expression level of APRT was high for more than 36 h which indicated that synthesized AHX might be promptly metabolized or converted to other derivative(s). Alternatively, AICAR might have been utilized for biosynthesis of more fundamental metabolites like IMP, inosine, hypoxanthine, uric acid and so on. In fact, AICAR formyltransferase (AICARFT), which catalyzes transfer of a formyl group to AICAR to produce N-formyl-5-aminoimidazole-4-carboxamide (FAICAR), was coded in the genome assembly of L. sordida37. Although AICARFT consumes AICAR and biosynthesizes the downstream metabolites, our data suggested that significant changes in expression level of AICARFT gene after AICAR feeding were not observed. The reason for this disagreement between results of feeding experiment and gene expression analysis in this study still remains unknown. Further analysis is needed to disclose other unknown metabolite(s) and derivative(s) in the future.
The consistent relationship between AICAR feeding and response of APRT at 12 h after feeding was observed in this report. Hence, what’s upstream of the APRT in L. sordida? To our knowledge, there is no report for transcriptional regulation mechanism of APRT in mushrooms. Mummaneni et al. (1993) reported that the promoter region of a mouse APRT was subjected to DNA methylation38. In the Chinese hamster, two putative GC factor (GCF) -binding sites suppress transcription of APRT39. In order to test a potential involvement of GCF for AHX biosynthesis and fairy ring formation, the feeding experiment of AICAR and the genome sequence in L. sordida would be useful. Detailed genome annotation of L. sordida is now underway to clarify further understanding of fairy ring formation.
Fungal genomes were analyzed by next-generation sequencing40. As fairy ring forming fungi, only P. involutus was sequenced41. However, there is no mechanistic information on the fairy ring formation in the species. Here, to promote elucidation of molecules for fairy ring formation mechanism, we conducted genome sequencing in L. sordida as its fairy ring formation mechanism was previously reported6. Hybrid assembly of the generated sequence reads provided a reference genome scaffold in L. sordida, and it was sufficient for identification of the APRT gene. The comparison of APRT sequences between L. sordida and P. involutus revealed conserved amino acid residues between fairy ring-forming fungi. The genomic information of L. sordida and P. involutus opens new avenues for elucidation of the mechanism of fairy ring formation.
A crystal structure analysis revealed the adenine binding domain of APRT in L. donovani23. This study reported Arg-40, Phe-41, and Ala-42 (corresponding to Val-22, Phe-23, and Leu-24, respectively, in L. sordida, Fig. 4) were residues to form hydrogen bonds with adenine of AMP. Interestingly, Sarver and Wang (2002) documented that a point mutation of the Phe of APRT in Giardia lamblia to Trp residue showed no obvious effect on adenine binding property42. Furthermore, the remaining two residues, Arg-40, and Ala-42 were substituted to Val and Leu, respectively, in mushroom-forming fungi (Fig. 4) and the substitutions caused a conformational change in region in our homology modeling (Fig. 6A). In L. sordida, the position of the Val residue shifted to just below Arg-40 of L. donovani APRT, and it extended the distance between adenine and the carbonyl group of the Val for the hydrogen bond (Fig. 6A and B). We propose the binding model of AICAR and AMP to L. donovani and L. sordida APRT as shown in Fig. 6B–D and E. The result suggested that L. sordida APRT has lower affinity to AMP, and might cause an increase in relative affinity to AICAR. Thus, decrease in affinity for AMP might affect the efficiency of AICAR usage in APRT from L. sordida.
Structure comparison between L. sordida and L. donovani APRT.
(A) Three-dimensional structural modeling was performed on SwissModel server with the L. donovani (PDB code, 1QB7) as the template. Ribbon diagram show the overlap of the L. donovani APRT monomer (blue) and L. sordida APRT monomer (yellow). Adenine, amino acid residues involved in the adenine binding of L. sordida (22-Val-Phe-Leu-24) and L. donovani (40-Arg-Phe-Ala-42) APRT are shown in red, green and orange, respectively. Distance between adenine and the carbonyl group is in angstroms. Schematic representation of putative adenine binding site of L. donovani APRT contacts to AMP (B) and AICAR (C). Schematic representation of putative adenine binding site of L. sordida APRT contacts to AMP (D) and AICAR (E).
Several other sites showing diversity between fairy ring-forming fungi and other organisms were found (Fig. 4). Although the position of 116 was neutral and acidic amino acid in other APRTs, Glu-116 was conserved between L. sordida and P. involutes (Fig. 4). Amino acid positioned on the dimer surface (due to electrical charge) might be involved resulting in their formation and the stability (Supplementary Fig. 3C). The position 139 was also substituted with Arg in fairy ring-forming fungi, and Arg-139 is component of α-helix near 5′ phosphate binding motif, 133-Ala-Thr-Gly-Gly-Ser-137 (Supplementary Fig. 3D). Although the side chain of Arg-139 was not facing the direction of the active site, its strong basicity might affect the conformation of active site. In the amino acid positions of 12, 34, 48, 53, 100, 165 and 167, residues were not conserved between fairy-ring forming fungi and other organisms (Fig. 4). In fairy ring-forming fungi, the position 53 in the missing region was substituted with a polar amino acid (Thr-53 in L. sordida or Arg-53 in P. involutes). The amino acid positioned at residue 165 was located above the active-site and might be involved in the attraction of the substrate.
In conclusion, we provided evidence of AHX biosynthetic pathway in L. sordida; 1) feeding of AICAR in L. sordida mycelia culture increased AHX accumulation and gene expression level of APRT, 2) APRTs among mushroom-forming fungi had evolutionarily conserved domains in active site, 3) L. sordida mycelia had an enzyme activity for conversion of AICAR to AICA. It will take some time to mine all genes involved in the biosynthesis of AHX and AOH. However, this study disclosed further metabolism of AICA which is a common member of the purine metabolic pathway in animals, plants, and microorganisms, whose metabolism had remained elusive.
Methods
Strain and culture condition
Lepista sordida provided from Nasu Biofarm, Ltd. (Japan) was pre-cultured in PYG medium (0.3% polypepton, 0.3% yeast extract, 1% glucose) supplemented with 1.8% of agar to obtain actively growing mycelia. The inoculated mycelia were incubated at 25 °C for 3 weeks under dark condition. After growth, the strain on the medium was stored at 4 °C. The mycelia were used within two months for following experiments below.
For preparation of genomic DNA, the strain was grown in YG medium (1% glucose, 0.3% yeast extract, 3.7 mM KH2PO4 and 3.5 mM Na2HPO4) in a flask with 120 rpm in dark condition. The culture was performed at 25 °C for 2 weeks. The mycelia were collected by filtration through 0.2 μm-membrane filter (Advantec Toyo Co., Japan).
For the feeding experiment and gene expression analysis, 8.5-mm-diameter of gel disks were punched out from the growing edge of mycelium. Two disks were each placed into a 50 mL Erlenmeyer flask containing 10 mL of YG medium. After incubation of the flasks at 25 °C for 12 days, 100 μL of 10 mM AICAR was added to the cultures, and then further incubated for 12, 24, 36, and 48 h. Cultured mycelia was prepared with five biological replicates.
RP-HPLC analysis
Amounts of AICAR and AHX in mycelia culture were determined by reversed-phase high-performance liquid chromatography (RP-HPLC). After feeding of AICAR to mycelia culture, 20 mL of methanol and 100 μL of 10 mM allopurinol (internal standard, final concentration 100 μM) were added to all of the samples. Then the cultures were homogenized by Polytron PT 1200E homogenizer (Kinematica, Switzerland) for 1 min. The cell suspensions were extracted with 20 mL of MeOH and filtrated. Each MeOH extract was dried at 35 °C by a rotary evaporator and re-dissolved in 10 mL of 10 mM ammonium formate buffer (pH 4.0), and then subjected to RP-HPLC. RP-HPLC was carried out to quantify AICAR and AHX under the following conditions: a PU-2089 gradient pump, AS-2010 multi wavelength detector, AS-2055 plus autosampler (JASCO Co., Japan); column, CAPCELL PAK ADME (4.6 mm × 250 mm; Shiseido Co., Japan); mobile phase, solution A: acetonitrile, solution B: 10 mM ammonium formate buffer (pH 4.0), 0–10 min, 2% Solution A; 10–40 min, 2–50% sol. A; flow rate,1 mL/min; and UV wavelength, 250 nm.
Detection of AICA by LC-MS/MS Analysis
AICAR-converting activity to AICA in crude enzyme extract of the mycelia was tested by LC-MS/MS analysis. The mycelia were frozen in liquid nitrogen and ground to a powder with a motor and pestle. The mycelia powder was extracted with extraction buffer containing 100 mM Tris-HCl (pH 8.3), 0.2% (w/v) 3-(3-cholamidepropyl) dimethylammonio-1-propanesulphonate, 10% (v/v) glycerol, 1 mM phenylmethylsulfonyl fluoride and 20 mM 2- mercaptoethanol at 4 °C for 3 h with shaking. The extract was centrifuged at 10,000 g for 20 min, and the supernatant was desalted by Centrifugal ultrafiltration using Amicon Ultra Centrifugal Filter Devices (3,000-Da cutoff; MerkMillipore, Japan). Ultrafiltration was repeated 3 times by adding fresh 100 mM Tris–HCl buffer (pH 8.3). After adding AICAR (final concentration 0.2 mM) to desalted crude extract, the sample was incubated at 28 °C for 8 h and subjected to LC-MS/MS analysis. AICA in the sample was detected on LC-MS/MS by comparing the retention time and precursor ion with those of authentic one. Analyses were performed with a Shimadzu UPLC system (Shimadzu, Japan) coupled to an LTQ Orbitrap mass spectrometer (Thermo Fisher Scientific, Waltham, MA, USA) equipped with an electrospray ionization probe. A PC-HILIC column (ϕ 2 × 100 mm, 3 μm; Shiseido, Japan) was used in the analysis (injection volume, 10 μL; solvent, 95% MeOH with 0.05% formic acid; flow rate; 0.2 mL/min). After each analysis, the column was re-equilibrated for 10 min at the initial conditions prior to the next sample analysis. MS analysis was performed in the negative FTMS mode at a resolution of 30,000 at m/z 400 with the following source parameters: sheath gas flow, 50; auxiliary gas flow rate, 10; tube lens, −63 V; capillary voltage, −16 V; ion spray voltage, 3 kV.
DNA preparation
The mycelia were frozen in liquid nitrogen and powdered with a motor and pestle. The powdered sample was used for extracting genomic DNA by using DNeasy Plant Mini Kit (Qiagen, Germany) following the instruction manual. Obtained DNA was quantified by using PicoGreen dsDNA Quantification Reagent (Invitrogen, USA) and analyzed on 0.7% of agarose electrophoresis to check the quality. The collected genomic DNA was used for genomic library construction.
Sequencing by Illumina/GAIIx
Two genomic DNA libraries for paired-end (PE) sequencing and mate paired-end (MP) sequencing were constructed by using Truseq DNA sample preparation kit and Mate pair library sample preparation kit, respectively, following the instruction manuals (Illumina, USA). For PE sequencing, 1 μg of genomic DNA was fragmented and electrophoresized on 2% of agarose to collect a DNA fraction of approx. 300 bp length. For MP sequencing, 10 μg of genomic DNA was fragmented and conducted to end-repaired. Subsequently, the 3′ ends of the DNA fragments were biotinylated by incorporating labeled dNTPs and electrophoresized on 0.6% of certified megabase agarose gel (Bio-Rad Laboratories Inc, USA) to collect a DNA fraction of approx. 3 kb length. DNA fragments in the fraction were circulated and then fragmented into 350–650 bp length. The biotinylated DNA was purified by using Dynabeads M-280 Streptavidin Magnetic Beads (Life Technologies, USA).
Each genomic DNA library was made blunt ended with End Repair Enzyme (Illumina) in the presence of 2.5 mM dNTPs and 10 mM ATP. Adenine nucleotide was added to the 3′ ends of the blunt ended cDNA with Klenow fragment (3′ to 5′ exominus) in the presence of 1 mM dATP by incubating at 37 °C for 30 minutes. The DNA with adenine on its ends was ligated with adapters provided by Illumina (PE; TruSeq Universal Adapter and TruSeq Indexed Adapter, MP; paired end (PE) oligo adapters) using T4 DNA ligase at room temperature for 15 min. The DNA was amplified with two adapter primers (Illumina) (PE; TruSeq Universal Adapter primer, and IlluminaTruSeq Adapter primer, MP; PE primers 1.0 and 2.0) with initial denaturing step at 98 °C for 30 s, followed by 15 cycles at 98 °C for 10 s, 65 °C for 30 s, 72 °C for 30 s with a final extension cycle at 72 °C for 5 min. The PCR products were purified with Qiaquick PCR purification kit and gel-extracted.
Whole genome sequencing using Illumina Genome Analyzer GAIIx (Illumina) was carried out by the method of 100 bp of paired-end. One lane for each library was used.
Sequencing by Roche/GS FLX Titanium
A paired-end genomic DNA library 8 K and a fragment genomic DNA library were constructed to be sequenced by GS FLX Titanium in Dragon Genomics Center (Takara Bio Inc., Japan). Construction of the paired-end library was conducted following the method described in GS FLX Paired End DNA Library Preparation Method Manual (provided from Roche Diagnostics K. K. Japan). Ten μg of the genomic DNA was mechanically digested by HydroShear (Digilab, USA) into approx. 8 kb of DNA fragments. The both termini of the DNAs were blunted and ligated with an adaptor with biotinated loxP site for cyclizing them. The DNA fraction was separated by agarose electrophoresis, and DNA fragments of 6.5 to 9.5 kb were eluted from the agarose gel by using the Elutrap Electroelution System (GE Healthcare, Japan). Obtained DNAs were circulated by a reaction of Cre Recombinase. The DNAs were then conducted to fragmentation by an Acoustic Solubilizer (Covaris, USA) and blunting the termini. After binding the DNAs to streptavidin-coupled dynabeads by using the affinity with biotin, the collected DNAs were ligated with the Library Adapters provided by the manufacture (Roche Diagnostics K. K.). The DNAs were amplified by PCR at 20 cycles by using a biotinated Amplification Primers (Roche Diagnostics K. K.) to purify the biotinated PCR products. Pool of denatured PCR products in alkaline treatment was designated as the paired-end library. For the fragment genomic DNA library, GS FLX Titanium Rapid Library Preparation Kit (Roche Diagnostics K. K.) was used following the manufacture’s manual. Briefly, one μg of the genomic DNAs was fragmentized by using Covaris S-series (Covaris, Inc., USA) and then ligated with RL adaptor included in GS FLX Titanium Rapid Library Preparation kit (Roche Diagnostics K. K.). From the DNA fraction, short DNAs were excluded by using Agencourt AMPure XP (Beckman Coulter, Inc., Brea, USA), and the resultant was designated as the sequence library.
Each sequence library was conducted to emulsion PCR after binding with capture beads. The capture beads were applied to sequencing by Genome Sequencer FLX + System.
Preprocessing of sequence reads
Raw sequence data of the Illumina sequence data were subjected to trimming and quality filtering were done with an in house Perl script. Low-quality bases at the sequence read ends with the quality value < 10 in fastq files were trimmed. Low-quality reads with either of the following criteria, read length < 20 bp, average of quality score per read < 17, number of low-quality bases (quality value < 10) per read > 10% or N’s per read > 0%, were removed.
Raw sequence data of the GS FLX sequence were processed by using the sff_extract software 0.3.0 (http://bioinf.comav.upv.es/sff_extract/index.html) to trim the adapter sequences.
De novo assembly
Sequence reads derived from GAIIx and GS FLX Titanium were conducted to hybrid assembly by multi-step procedures. The sequence reads from GS FLX Titanium were assembled into contigs by Newbler43, and then the contigs were further integrated with Illumina mate pair short reads to generate genomic scaffolds by SSPACE44. At final, Illumina pair end reads were used to fill an indefinite nucleotide “N” in the genomic scaffolds by Gap closer45.
Sequence search
HMMER (ver. 3.1) was applied to search an APRT gene sequence in L. sordida46. Query sequence was conducted to nhmmer search against the whole genome assembly in L. sordida with the default setting.
cDNA cloning of APRT in L. sordida
RT-PCR was performed with PrimeScriptTM RT-PCR kit (TaKaRa Bio Inc., Japan). Total RNA was prepared by using RNeasy plant mini kit (Qiagen) and applied to 1st-stranded cDNA synthesis with oligo (dT) primer and a PrimeScript RT reagent kit (TaKaRa Bio Inc.). A pair of primers (5′-ATGGACGTTGAGTACATTAAAG-3′ and 5′-TCAATCATCCGATTGAACGATC-3′) were designed based on predicted nucleotide sequence of APRT gene and employed for PCR. Amplified PCR product was purified and cloned in a vector pMD20-T (TaKaRa Bio Inc.) using a Mighty TA cloning kit (TaKaRa Bio Inc.).
Structural analysis of APRT protein
Multiple alignment of APRT was prepared using the ClustalX software (Thompson et al., 1997). The secondary structure of the L. sordida APRT was predicted by PSIPRED v3.3 on the PSIPRED server47. We predicted a protein structure of APRT in L. sordida using the Swiss-Model automated protein structure homology-modeling server with the template of the APRT in Saccharomyces cerevisiae (Protein data bank code, 1G2Q)48. Individual hydrophobicity was determined using the Protscale software at the EXPASY server (Gasteiger et al. 2005), with the amino acids scale of “Hphob/Kyte and Doolittle”49. Distance between adenine and the carbonyl group in the homology model was calculated with Pymol (https://www.pymol.org/).
Quantitative gene expression analysis
Gene expression analysis was carried out using quantitative real-time RT-PCR (qRT-RT-PCR). qRT-RT-PCR was performed in 96-well plates with a LightCycler 480 real-time PCR instrument (Roche Diagnostics K. K.). One microgram of total RNA samples was incubated with 1 U deoxyribonuclease I (Invitrogen, USA) for 15 min at room temperature to remove genomic DNA, and EDTA was added to a final concentration of 2 mM to stop the reaction. After that, 1st-strand cDNAs were synthesized from total RNAs using a PrimeScript RT reagent kit (TaKaRa Bio Inc.) with oligo (dT) primer. Each 20 μL reaction mixture containing 50 ng of total cDNA, 10 μL FastStart Essential DNA Green Master (Roche Diagnostics K. K.), and 0.5 μM of each primer. Cycling conditions was set as follows: pre-incubation, 1 cycle of 95 °C for 10 min; amplification, 45 cycles of 95 °C for 10 s, 57 °C for 10 s and 72 °C for 10 s. The following primers were used: APRT forward primer, 5′-CTCCTCGGTCCAATAATCGC-3′; APRT reverse primer, 5′-AAATATGTCCACACCGTATTC-3′; AICARFT forward primer, 5′-CAAGATTGACGCTAAGCTCTTCGAG-3′; AICARFT reverse primer, 5′- GTGGCGACGATTAAGTCTGTAATTG. Gene expression levels were normalized by gene expression of an actin; primers 5′-CGTTGTTTGCGGTCG-3′ and 5′-GCTAAAATATCTTTG-3′ were designed.
Additional Information
How to cite this article: Suzuki, T. et al. The biosynthetic pathway of 2-azahypoxanthine in fairy-ring forming fungus. Sci. Rep. 6, 39087; doi: 10.1038/srep39087 (2016).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Couch, H. B. Diseases of turfgrasses. Krieger, Malabar 181–186 (1995).
Smiley, R. W., Dernoeden, P. H. & Clarke, B. B. Compendium of turfgrass diseases. APS Press, Minneapolis, MN, 98 (1992).
Smith, J. D., Jackson, N. & Woolhouse, A. R. Fungal diseases of amenity turf grasses. E.&F. N. Spon, London 339–353 (1989).
Shantz, H. L. & Piemeisel, R. L. Fungus fairy rings in eastern Colorado and their effect on vegetation. J. Agric. Res. 191–245 (1917).
Terashima, Y. & Fujiie, A. Comparison of conditions for mycelial growth of Lepista sordida causing fairy rings on Zoysia matrella turf to those on Agrostis palustris turf. Mycosci. 48, 365 (2007).
Choi, J. H. et al. Disclosure of the “fairy” of fairy-ring-forming fungus Lepista sordida. Chembiochem 11, 1373–1377 (2010).
Choi, J. H. et al. The source of “fairy rings”: 2-azahypoxanthine and its metabolite found in a novel purine metabolic pathway in plants. Angew. Chem. Int. Ed. 53, 1552–1555 (2014).
Tobina, H. et al. 2-Azahypoxanthine and Imidazole-4-carboxamide produced by the fairy-ring-forming fungus increase yields of wheat. Field Crop Res. 162, 6–11 (2014).
Shi, W. et al. Structural analysis of adenine phosphoribosyltransferase from Saccharomyces cerevisiae. Biochemistry 40, 10800–10809 (2001).
Krenitsky, T. A., Neil, S. M., Elion, G. B. & Hitchings, G. H. Adenine phosphoribosyltransferase from monkey liver. Specificity and properties. J. Biol. Chem. 244, 4779–4784 (1969).
Flaks, J. G., Erwin, M. J. & Buchanan, J. M. Biosynthesis of the purines XVI. The synthesis of adenosine 5′-phosphate and 5-amino-4-imidazolecarboxamide ribotide by a nucleotide pyrophosphorylase. J. Biol. Chem. 228, 201–213 (1957).
Sin, I. L. & Finch, L. R. Adenine phosphoribosyltransferase in Mycoplasma mycoides and Escherichia coli. J. Bacteriol. 112, 439–444 (1972).
Chen, C. M., Melitz, D. K. & Clough, F. W. Metabolism of cytokinin: phosphoribosylation of cytokinin bases by adenine phosphoribosyltransferase from wheat germ. Arch. Biochem. Biophys. 214, 634–641 (1982).
Dush, M. K., Sikela, J. M., Khan, S. A., Tischfield, J. A. & Stambrook, P. J. Nucleotide sequence and organization of the mouse adenine phosphoribosyltransferase gene: presence of a coding region common to animal and bacterial phosphoribosyltransferases that has a variable intron/exon arrangement. Proc. Nat. Acad. Sci. 82, 2731–2735 (1985).
Hidaka, Y., Tarle, S. A., O’Toole, T. E., Kelley, W. N. & Palella, T. D. Nucleotide sequence of the human APRT gene. Nucleic Acids Res. 15, 9086 (1987).
Moffatt, B. A., McWhinnie, E. A., Agarwal, S. K. & Schaff, D. A. The adenine phosphoribosyltransferase-encoding gene of Arabidopsis thaliana. Gene 143, 211–216 (1994).
Itai, R. et al. Induced activity of adenine phosphoribosyltransferase (APRT) in iron-deficiency barley roots: a possible role for phytosiderophore production. J. Exp. Bot. 51, 1179–1188 (2000).
Hammond, D. J. & Gutteridge, W. E. Purine and pyrimidine metabolism in the Trypanosomatidae. Molecular and biochemical parasitology 13, 243–261 (1984).
Myers, R. W. & Abeles, R. H. Conversion of 5-S-ethyl-5-thio-D-ribose to ethionine in Klebsiella pneumoniae. Basis for the selective toxicity of 5-S-ethyl-5-thio-D-ribose. J. Biol. Chem. 264, 10547–10551 (1989).
Bollee, G. et al. Adenine phosphoribosyltransferase deficiency. Clin. J. Am. Soc. Nephrol. 7, 1521–1527 (2012).
Moffatt, B. & Somerville, C. Positive selection for male-sterile mutants of Arabidopsis lacking adenine phosphoribosyl transferase activity. Plant Physiol. 86, 1150–1154 (1988).
Chan, C. H. et al. Dissecting cobamide diversity through structural and functional analyses of the base-activating CobT enzyme of Salmonella enterica. Biochim. Biophys. Acta 1840, 464–475 (2014).
Phillips, C. L., Ullman, B., Brennan, R. G. & Hill, C. P. Crystal structures of adenine phosphoribosyltransferase from Leishmania donovani. EMBO J. 18, 3533–3545 (1999).
Silva, M., Silva, C. H., Iulek, J., Oliva, G. & Thiemann, O. H. Crystal structure of adenine phosphoribosyltransferase from Leishmania tarentolae: potential implications for APRT catalytic mechanism. Biochim. Biophys. Acta 1696, 31–39 (2004).
Silva, M., Silva, C. H., Iulek, J. & Thiemann, O. H. Three-dimensional structure of human adenine phosphoribosyltransferase and its relation to DHA-urolithiasis. Biochemistry 43, 7663–7671 (2004).
Shi, W. et al. Closed site complexes of adenine phosphoribosyltransferase from Giardia lamblia reveal a mechanism of ribosyl migration. J. Biol. Chem. 277, 39981–39988 (2002).
Jensen, K. F. et al. Adenine phosphoribosyltransferase from Sulfolobus solfataricus Is an enzyme with unusual kinetic properties and a crystal structure that suggests It evolved from a 6-oxopurine phosphoribosyltransferase. Biochemistry 54, 2323–2334 (2015).
Johnson, D. H. Adenine phosphoribosyltransferase genes in two Drosophila species: dosage compensation, a nuclear matrix attachment site, and a novel intron position. Mol. Gen. Genet. 238, 383–389 (1993).
Fieldhouse, D. & Golding, G. B. The rat adenine phosphoribosyltransferase sequence shows evolutionary rate variation among exons in rodents. Genome/National Research Council Canada = Genome/Conseil national de recherches Canada 36, 1107–1110 (1993).
Schnorr, K. M., Gaillard, C., Biget, E., Nygaard, P. & Laloue, M. A second form of adenine phosphoribosyltransferase in Arabidopsis thaliana with relative specificity towards cytokinins. Plant J. 9, 891–898 (1996).
Vos, S., de Jersey, J. & Martin, J. L. Crystal structure of Escherichia coli xanthine phosphoribosyltransferase. Biochemistry 36, 4125–4134 (1997).
Ozturk, D. H., Dorfman, R. H., Scapin, G., Sacchettini, J. C. & Grubmeyer, C. Structure and function of Salmonella typhimurium orotate phosphoribosyltransferase: protein complementation reveals shared active sites. Biochemistry 34, 10764–10770 (1995).
Scapin, G., Grubmeyer, C. & Sacchettini, J. C. Crystal structure of orotate phosphoribosyltransferase. Biochemistry 33, 1287–1294 (1994).
Schumacher, M. A., Carter, D., Roos, D. S., Ullman, B. & Brennan, R. G. Crystal structures of Toxoplasma gondii HGXPRTase reveal the catalytic role of a long flexible loop. Nat. Struct. Biol. 3, 881–887 (1996).
Krahn, J. M. et al. Coupled formation of an amidotransferase interdomain ammonia channel and a phosphoribosyltransferase active site. Biochemistry 36, 11061–11068 (1997).
Focia, P. J., Craig, S. P., 3rd & Eakin, A. E. Approaching the transition state in the crystal structure of a phosphoribosyltransferase. Biochemistry 37, 17120–17127 (1998).
Hartman, S. C. & Buchanan, J. M. Biosynthesis of the purines XXVI. The identification of the formyl donors of the transformylation reactions. J. Biol. Chem. 234, 1812–1816 (1959).
Mummaneni, P., Bishop, P. L. & Turker, M. S. A cis-acting element accounts for a conserved methylation pattern upstream of the mouse adenine phosphoribosyltransferase gene. J. Biol. Chem. 268, 552–558 (1993).
She, B. R. & Taylor, M. W. Identification of the cis-elements required for transcriptional control of the Chinese hamster ovary APRT gene. Gene 159, 175–180 (1995).
Grigoriev, I. V. et al. The genome portal of the Department of Energy Joint Genome Institute. Nucleic Acids Res. 40, D26–32 (2011).
Kohler, A. et al. Convergent losses of decay mechanisms and rapid turnover of symbiosis genes in mycorrhizal mutualists. Nat. Genet. 47, 410–415 (2015).
Sarver, A. E. & Wang, C. C. The adenine phosphoribosyltransferase from Giardia lamblia has a unique reaction mechanism and unusual substrate binding properties. J. Biol. Chem. 277, 39973–39980 (2002).
Margulies, M. et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437, 376–380 (2005).
Boetzer, M., Henkel, C. V., Jansen, H. J., Butler, D. & Pirovano, W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics (Oxford, England) 27, 578–579 (2011).
Luo, R. et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience 1, 18 (2012).
Wheeler, T. J. & Eddy, S. R. Nhmmer: DNA homology search with profile HMMs. Bioinformatics (Oxford, England) 29, 2487–2489 (2013).
Buchan, D. W., Minneci, F., Nugent, T. C., Bryson, K. & Jones, D. T. Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Res. 41, W349–357 (2013).
Biasini, M. et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42, W252–258 (2014).
Kyte, J. & Doolittle, R. F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157, 105–132 (1982).
Acknowledgements
This work was partially supported by a Grant-in-Aid for the Programme for Promotion of Basic and Applied Researches for Innovations in Bio-oriented Industry and a Grant-in-Aid for Scientific Research on Innovative Areas “Chemical Biology of Natural Products” from the Ministry of Education, Culture, Sports, Science and Technology, Japan (MEXT) to H. Kawagishi. This work was also supported in part by Grants in-Aid for Scientific Research on Innovative Areas (No. 24113518 and No. 26113716 to K.Yano) from MEXT and the MEXT-Supported Program for the Strategic Research Foundation at Private Universities (2014–2018) and Research Funding for the Computational Software Supporting Program from Meiji University to K. Yano. Computations were partially performed on the NIG supercomputer at ROIS National Institute of Genetics. We thank Dr. V. K. Deo (Shizuoka University) for valuable discussion.
Author information
Authors and Affiliations
Contributions
H.K. conceived the project and designed outlines most of the experiments. K.Y. designed the bioinformatics approaches. T.S., J.-H.C. and Y.T. performed feeding experiments. H.D. and H.H. designed PCR experiments. T.S.Y.T. and A.I. performed PCR experiments. T.S., N.Y., T.T., Y.S. and Y.N. performed genome sequencing and analysis of the resulting data. T.S. and N.Y. performed other experiments and analyzed data. T.S., N.Y. and H.K. wrote the manuscript. All authors reviewed the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Suzuki, T., Yamamoto, N., Choi, JH. et al. The biosynthetic pathway of 2-azahypoxanthine in fairy-ring forming fungus. Sci Rep 6, 39087 (2016). https://doi.org/10.1038/srep39087
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep39087
This article is cited by
-
Assessing soil microbes that drive fairy ring patterns in temperate semiarid grasslands
BMC Ecology and Evolution (2022)
-
Grassland fairy rings of Leucocalocybe mongolica represent the center of a rich soil microbial community
Brazilian Journal of Microbiology (2021)
-
Genome sequence analysis of the fairy ring-forming fungus Lepista sordida and gene candidates for interaction with plants
Scientific Reports (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.