Abstract
The Sunda Arc forms an almost continuous chain of islands and thus a potential dispersal corridor between mainland Southeast Asia and Melanesia. However, the Sunda Islands have rather different geological histories, which might have had an important impact on actual dispersal routes and community assembly. Here, we reveal the biogeographical history of hyperdiverse and flightless Trigonopterus weevils. Different approaches to ancestral area reconstruction suggest a complex east to west range expansion. Out of New Guinea, Trigonopterus repeatedly reached the Moluccas and Sulawesi transgressing Lydekker′s Line. Sulawesi repeatedly acted as colonization hub for different segments of the Sunda Arc. West Java, East Java and Bali are recognized as distinct biogeographic areas. The timing and diversification of species largely coincides with the geological chronology of island emergence. Colonization was not inhibited by traditional biogeographical boundaries such as Wallace’s Line. Rather, colonization patterns support distance dependent dispersal and island age limiting dispersal.
Similar content being viewed by others
Introduction
The Indo Australian Archipelago (IAA) is the geologically complex transition zone between Oriental and Australian biota and arguably the most biodiverse region on the planet1. Biogeographers ever since Wallace have sought to understand the mechanisms behind the high diversity of the IAA and its evolution in space and time. Recently, several comprehensively sampled molecular phylogenies have helped to reveal underlying historical processes e.g. across Sundaland2, around Sulawesi3, the Philippines4,5 and Melanesia6. However, the Sunda Arc along the southern border of the IAA7 remains poorly studied, especially so the Lesser Sunda Islands comprising Bali, Lombok, Sumbawa and Flores. The pioneering work of Rensch8 reflects a great interest at the time but became outdated with the advent of plate tectonic theory and phylogenetics9,10.
Yet, the Sunda Arc provides a compelling setting to study biotic evolution along a complex island chain. Its islands are arranged like pearls on a string, slightly south of the equator. The waterways between islands are usually narrow, e.g. Bali Strait between Java and Bali is currently less than 3 kilometres wide, or Alas Strait between Lombok and Sumbawa <15 km. Today and for approximately the last ten million years the islands of the Sunda Arc were aligned along the same trench, but their age and geological history differ greatly11,12. While Java is geologically of continental origin, the volcanic influence markedly increases towards its East. Bali, Lombok and Sumbawa are entirely of volcanic origin, emergent since ~11 million years ago (Ma) while Flores to the east presumably emerged later, c. 9 Ma12. Sumba is a remarkable exception, being a continental splinter from the Sunda Shelf pushed to the southeast13.
During the Pleistocene glacial maxima those islands located on the Sunda Shelf were connected with each other and the Asian mainland14,15. The resulting faunal exchange, e.g. between Java and Bali resulted in faunal similarities which are in stark contrast to the ones of islands further east. The deepwater Lombok Strait between Bali and Lombok, however, was never dry14,16. The resulting differences particularly in vertebrate faunas to the east and west of the Lombok Strait led Wallace17 to suggest a sharp division of biogeographical regions, known as Wallace’s Line16,18.
Habitats in the exposed Java Sea connecting the Sunda Shelf islands might have included savannah, heath forests or peat swamps19. These acted as an ecological filter favouring taxa of higher mobility, particularly mammals20,21,22,23. Some species, e.g. the tiger (Panthera tigris Linnaeus) had reached Sumatra, Java and Bali but were always absent from Borneo24. This Pleistocene faunal exchange occurred only in the relatively recent past of a much older fauna. The story of Wallace’s Line thus appears relevant only for some taxa and for just a fraction of the time-span during the biotic evolution of the IAA. The climatic processes producing the high visibility and prominence of Wallace’s Line may in fact distract from older geological patterns of faunal assembly in the IAA25.
Arthropods constitute the vast majority of species diversity across the IAA and the Sunda Arc, but their biogeographic origins remained mostly unknown until recently21,26,27,28,29,30,31,32,33,34. In some cases they show unexpected colonization histories including long-distance dispersal35,36. Some of the taxa with broad distributions across the IAA have a natural history that appears to be counterintuitive to the observed patterns, i.e. they are flightless and/or have little tolerance to seawater. Such taxa include the fanged frogs that originated in SE Asia and then spread across the IAA, with frequent exchange between Borneo, Philippines and Sulawesi, crossing both Wallace’s and Huxley’s lines37. Flightless Trigonopterus weevils crossed Wallace’s Line three times to reach Bali from the east38, mite harvestmen reached New Guinea twice from Sundaland26 and land snails reached the Lesser Sunda Islands from Australia39.
Here we study hyperdiverse and flightless Trigonopterus weevils that might have more than 1,000 species across the region40,41. This extraordinary diversity, limited dispersal ability and high degree of local endemism allows for fine scale analyses of patterns and processes in the faunal evolution of the IAA and the Sunda Arc. The genus Trigonopterus has a wide range, from east Sumatra across Melanesia to Samoa. New Guinea is a centre of diversity (>300 species recorded,42). Sulawesi is another hotspot with >100 species (Riedel, in prep.). Diversity decreases to the west, but is still substantial compared to other organisms; there are at least 100 species in Sundaland and the Lesser Sunda Islands43. All except two species are microendemics. The high level of species endemism provides many replicated natural experiments to understand the processes generating tropical diversity. Here we explore their biogeographical history and colonization patterns of the IAA in general and the Sunda Arc in particular.
Trigonopterus of Sundaland and the Lesser Sunda Islands are exclusively edaphic, inhabiting the leaf-litter, while in Sulawesi and New Guinea many species browse on foliage. In the relatively dry islands of Sumbawa and Flores, Trigonopterus are confined to montane areas of wet primary forests while they are absent from seasonal monsoon forests. Extensive areas of East Java and the Lesser Sunda Islands at low elevations and/or in rain shadows do not support habitats suitable for Trigonopterus, perhaps similar to the exposed areas of the Sunda Shelf during the Pleistocene glacial maxima.
Based on comprehensive taxon sampling we reconstruct the molecular phylogeny for the Sunda Arc Trigonopterus, infer divergence time estimates using multiple calibration strategies and use these data to investigate the biogeographical history of this lineage. Our findings suggest that lineage accumulation over time largely coincides with relative island age except for Sumatra and Borneo both of which were presumably outside of the initial area of range expansion. We find multiple east-west range expansion events out of New Guinea. Sulawesi acted as a colonization hub for multiple independent colonization events across different segments of the Sunda Arc. The Sunda Arc’s fauna is the result of a complex over water colonization history, with no back colonization to source areas.
Results
Molecular phylogenetics
The best-fit partitioning strategy recovered by PartitionFinder is shown in Suppl.-Table 11. The Bayesian inference (BI) analyses converged well with ESS values >200 for all parameters. The Bayesian inference, maximum parsimony and maximum likelihood reconstructions recovered highly congruent topologies for the species of the Sunda Arc (Fig. 1, S1, S3). Node support was high (≥0.95 posterior probability in BI) for nodes critical to our inference (Fig. 1, S1). Some clades of the New Guinea/Moluccas have lower nodal support (e.g. clades D, E), but they are nested among other clades of the same areas and therefore do not affect our conclusions.
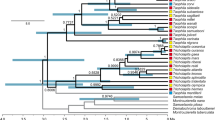
Historical biogeography of Trigonopterus weevils inferred from model-based analysis.
The Bayesian phylogeny was dated using the re-emergence of Java at 10 Ma (analysis 2). The chronogram is presenting the median divergence time estimates resulting from the BEAST analysis. Ancestral areas were inferred using a time-stratified DEC + J + x model. The distribution of each taxon is given in a geographical matrix on the right side of the chronogram with colours as coded in the inset. Coloured pie-charts indicate the likelihood of ancestral areas as recovered at each node. Nodes referred to in the text are marked by letters A to O. The map inset was created in GeoMapApp v3.4.1 http://www.geomapapp.org and modified in Adobe Photoshop CS2.
Divergence time estimation
Using the emergence and formation of a larger New Guinea landmass beginning at 30 Ma placed the root of Trigonopterus at 29.9 Ma with a 95% height posterior distribution (HPD) of 27.8–31.7 Ma (Analysis 1, Fig. S2). This suggests a CO1 substitution rate of 0.0315 substitutions per site per million year (Myr) per lineage (subs/s/Myr/l) (0.0342 subs/s/Ma/l for the 3′ fragment). Analysis 2 was based on the re-emergence of Java at 10 Ma following a complete inundation of shallow sea11,44. This recovered the root of Trigonopterus at 11.6 Ma (Fig. 1) (95% HPD: 9.1–14.2 Ma) resulting in CO1 substitution rates of 0.0793 subs/s/Myr/l (0.0865 subs/s/Myr/l for the 3′ fragment).
Biogeographical model testing
The results from the Maximum Likelihood (ML) estimation of biogeographical models are given in S-Table 8, which also provides the results for the model parameters d (rate for range expansion), e (rate for range contraction/extinction), J (allowing for jump dispersal) and x (a dispersal rate multiplier)45,46. The parameters J and x are fixed when reported as 0 values. The results of the Likelihood Ratio Tests (LRT) between models with and without the +J parameter (S-Table 9) are all in favour of the Dispersal-Extinction-Cladogenesis (DEC)-type models that included the +J parameter. Comparison of the different models with Akaike Information Criterion corrected for sample size (AICc) (Table 1), revealed support for the DEC + J + x model with dispersal constrained to adjacent areas except for the Lesser Sunda Islands chain. A constrained model receiving a higher log-likelihood (LnL) may seem counterintuitive. However, this is possible by giving a higher probability to just a few “correct events” (i.e. giving a better ML estimate with fewer events) to explain the tip data while low probability events are ruled out, thus optimizing d and e. The two constraint models received less support when distance was not included. The constrained model where dispersal was allowed between any of the Lesser Sunda Islands always received more support over the “adjacency only constrained model” where dispersal was only allowed between adjacent islands in the Lesser Sunda Islands chain. The unconstrained model received less support than the constrained model where dispersal was allowed between any of the Lesser Sunda Islands (Table 1).
Parsimony area reconstruction
The results from the parsimony area reconstruction (Fig. S1) were broadly congruent with the preferred BioGeoBEARS analyses using the DEC + J + x model (Fig. 1, S2). This demonstrates the robust signature of the following biogeographical history.
Sunda Islands biogeography in space
Trigonopterus of the Wallacea, the Philippines and Sundaland (see1 for a definition of these areas) originated in New Guinea from where they expanded their range westward repeatedly transgressing Lydekker’s and Wallace’s Line (Figs 1 and 2; S1). The Moluccas and Sulawesi were stepping stones to the Sunda Arc; Sulawesi also evolved numerous endemic species. There were at least 12 westward colonization events out of New Guinea (Fig. 1, S1 clades A–F; Fig. 2). The vast majority of species west of New Guinea belongs to clade G, which is also the oldest westward range expansion (Fig. 1).
Synopsis of major dispersal events of Trigonopterus.
From New Guinea, Trigonopterus repeatedly reached the Moluccas and Sulawesi transgressing Lydekker′s Line. Sulawesi became a secondary centre of diversification and colonization hub for different segments of the Sunda Arc with independent lineages. The map was created in GeoMapApp v3.4.1 http://www.geomapapp.org and modified in Adobe Photoshop CS2.
The early lineages of clade A are from New Guinea and the Moluccas from where Sulawesi and West Java were colonized independently; East Java and Sumatra were repeatedly colonized from West Java. Clade B originated in New Guinea from where it reached the Moluccas and Sulawesi and in an independent subclade Sulawesi and Sumbawa. Clades C, D and E reached the Moluccas from New Guinea. In clade F, we find a westward range expansion from New Guinea to Borneo and the Philippines. Clade G contains almost the entire fauna of Sulawesi, Borneo and the Sunda Arc, with four independent colonization events from Sulawesi towards the Sunda Arc followed by subsequent diversification: 1) clade H to East Java/Sumbawa and later to Bali, Lombok and Flores; 2) clade J to Flores and from there to the other Lesser Sunda Islands; 3) the common ancestor of clades L and M to Java; 4) clade O to Lombok/Sumbawa and from there to Flores, followed by two independent colonization events to Bali38.
In Java, we find an early dichotomy between West Java (clade M) and East Java (clade L). There is little exchange between the two areas in clade A. West Java acts as a launching pad to Sumatra in clades A and M. East Java is the stepping stone to the Lesser Sunda Islands for clade L. There is no exchange between West and East Java in the relatively diverse clades L and M.
Sunda Islands biogeography in time
Trigonopterus reached Sulawesi or parts thereof relatively early during their evolution, according to our tentative dating either 22.0 Ma using the emergence of New Guinea as calibration, or 8.0 Ma using the emergence of Java as a calibration (in the following, dates of this second calibration in parentheses) (Fig. 1, S2). The colonization of the Sunda Islands started with the ancestor of clades L and M in Java at 16.8 Ma (6.2 Ma) and continued with clade J in the Lesser Sunda Islands 15.6 Ma (5.6 Ma). With slight delay followed clade A in Java at 14 Ma (4.9 Ma), respectively clade O in the Lesser Sunda Islands at 13.6 Ma (4.7 Ma). Colonization of Sumatra started much later with lineages of clade M at 5.6 Ma (3.2 Ma). Colonization of Bali by lineages of clade O commenced at 7.0 Ma (1.5 Ma). The relatively recent radiation of clade N comprising eight species of mid to upper montane forests of West Java begins 5.0 MA (2.2 MA).
Discussion
Substitutions rates for Trigonopterus (Analysis 1, 2) were here inferred from large scale paleogeographical events. These rates were up to 7.5 times higher (0.0115 vs 0.0865) than other published arthropod rates ranging from 0.0115 to 0.0195 subs/s/Myr/l (e.g.47,48,49,50). This however is in line with recent evidence linking flight loss to accelerated molecular evolution (Mitterboeck & Adamowicz, 2013), which was suspected for flightless Trigonopterus as well38. Thus, the timing of events remains tentative in the absence of specific ingroup fossils. Yet, our ancestral area estimations were largely congruent among different analyses (undated parsimony versus differently dated model based). This may in part be due to each species being found in only one area and Trigonopterus of the IAA being strongly structured geographically. While the parsimony analysis is free of temporal assumptions, the sequence of events in the analysis reveals a relative chronological order, which agrees with the dated, model based ancestral area inferences.
Our results reveal a New Guinean origin of Wallacean and Sundaland Trigonopterus. Species diversity of the Sunda Arc Islands has been derived through at least six colonization events followed by in situ diversification on different segments of the Sunda Arc (Fig. 2, S1). The highly diverse clade G arrived and diversified in Sulawesi and then expanded west or south with four colonization events to the Sunda Arc each followed by rapid diversification. It remains west of the Moluccas, with no back colonization to New Guinea or the Moluccas. Thus, the overall directionality of the range expansion is in the east to west direction. Colonization of the Sunda Islands equally appears as a one-way street, at least when following the parsimony and the second model-based analysis respectively. The first model-based analysis interprets clade K as a back-colonization of Sulawesi from Java, but this scenario does not appear compelling, adding more steps to the colonization process.
Here, we show that the Sunda Arc is biogeographically broken into different segments, not all conforming to passages between islands or biogeographic lines such as Wallace’s. The Bali Strait is the most striking barrier, separating the continental island of Java from the volcanic Lesser Sunda Islands. Only two transgressions have occurred here between East Java and Lombok/Sumbawa (Fig. 1, S1, clade H, clade L), which is remarkable considering the rich faunas on either side not transgressing this barrier. In comparison, Wallace’s Line between Bali and Lombok and the Sunda Strait between Sumatra and Java were distinctly more porous with three and four transgressions respectively38. Apparently, the geological boundary between continental and volcanic islands is more significant than Wallace’s Line, largely inferred from climatic effects of Pleistocene sea level minima. A simple relationship between the age of terranes and the number of transgressions can be ruled out because most of these transgressions took place at a time when all islands were already available to colonization according to the Java calibrated reconstruction. However island age cannot be entirely ruled out as a significant factor contributing to the colonization process, as there is uncertainty in the age of the phylogeny.
This study also reveals an intra-island division between West and East Java with deep divergence of clades L and M (Fig. 1, S1). This pattern appears to confirm the separation of West and East Java as separate islands that re-emerged from the Java Sea between 10 to 5 Ma and coalescing to form Java in its current shape only quite recently11,44. The Javanese species of clade A (Fig. 1, S1) show two transgressions of Central Java, but these occurred at ca. 3.2 Ma and 4.3 Ma based on model-based analysis 2, at a time when West and East Java were possibly already connected. Dispersal among the Lesser Sunda Islands was relatively frequent, also reflected in the model-based analyses by the adjacency constraints receiving a lower likelihood than the unconstrained results.
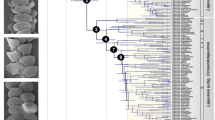
Sulawesi’s early colonization entailing prompt diversification (Fig. 3) and its central position in the heart of the IAA made it a launching pad for the colonization of the more recent Sunda Islands. From Sulawesi there are five colonisation events to the rest of the Sunda Arc, plus three to Borneo. This leaves only two long distance colonization events from New Guinea/Moluccas directly to the Sunda Islands (Fig. 1, S1, clades A, F), which might however be artefacts caused by missing species from interspersed areas.
Lineage accumulation per area over time, revealing range expansion as new resources become available or in reach.
Numbers of lineages are counted in time-slices of 2 MA-intervals in analysis 2 (see Fig. 1). For New Guinea, only a subset of the available species diversity was used resulting in negative bias for recent lineages.
Sulawesi was colonized at different time slices: clade G arrived ca. 22.0 Ma (8.0 Ma), clade B arrived 12.2 Ma (5.8 mya), respectively 2.0 Ma (1.0 Ma) and clade A 6.5 Ma (2.0 mya) (Analysis 1/2, respectively). This step-by-step accumulation of lineages over time (Fig. 3) rather than simultaneous arrival suggests a prominent role of over water dispersal rather than major vicariance events in the formation of the Sulawesi fauna. The earliest inferred arrival at 22.0 Ma coincides with the collision of the Sula spur, a large promontory of the Australian continental margin, with the volcanic arms of north/northeast Sulawesi12,51 and although this may not have transposed an entire fauna to proto-Sulawesi, it was probably a time promoting dispersal by the close contact of the respective terranes.
Thus, concerning the colonization of Sulawesi the time-frame of analysis 1 is consistent with current geological opinion. The almost instant diversification appears somewhat earlier than in other taxa, generally starting not before 20 Ma51 and attributed to the small area of proto-Sulawesi at its early days. However, it is more problematic to see colonization and subsequent radiation on Java at 18 Ma and at Flores at 15 Ma. Hall52 suggests that “most of Sumatra and Java were elevated above sea level and emerged to their present size only since 5 Ma and much of East Java continued to be the site of marine deposition until late in the Pliocene or even Pleistocene.” Flores appeared as the last of the Lesser Sunda Islands at 9 Ma12. Therefore, analysis 2 was performed which resulted in a staggering evolutionary rate of 0.0793 subs/s/Ma/l in CO1 but at the same time in an overall scenario more consistent with current geological opinion11,12,44,52. Here, Java would have been colonized by the first lineage at 6.2 Ma and Flores at 5.8 Ma. In the reconstructions of Spakman & Hall12 the islands of Bali, Lombok and Sumbawa appear at 11 Ma and Flores slightly later at 9 Ma although the latter was the first of the Lesser Sundas with a radiation of Trigonopterus. According to Barberi et al.53 Sumbawa may be largely of Pleistocene age with Mt. Sangenges dated as 1.71 Ma. This agrees well with the age of the T. sumbawensis-clade P found at this mountain and neighbouring Mt. Batu Pasak dated 1.5 Ma according to analysis 2. Similarly, according to analysis 2 clade N radiated largely during the Pleistocene concurring with Quarternary volcanism building mountains such as Mt. Gede7. Diversification in New Guinea would have commenced at 11.6 Ma, which would be contemporaneous with the second phase of orogeny in New Guinea, the uplift of the Central Range54. Opening of the Sunda Strait started before 2 Ma55, which is consistent with analysis 2 where all four dispersal events from Java to Sumatra took place shortly before this time. Since then the Sunda Strait has acted as an effective barrier for dispersal between West Java and Sumatra.
The colonization of Lombok (Fig. 1, S1, clades J, L, O) appears relatively early in time, compared to the colonization of Bali (Fig. 1, S1, clade O), a geologically similar island; however, recent volcanic deposits may have coved earlier rocks, making it very difficult to determine island age by stratigraphy. Possibly, our results could indicate a longer above-water-history of Lombok compared to Bali. Biogeographical data of other organisms should be examined for congruence. Unfortunately only a few published phylogenies include representatives of single island endemics for the Western Lesser Sunda Islands. For instance in37, where one species of fanged frogs (Limnonectes kardasani) an endemic to Lombok was included but the absolute age is not provided for its divergence between the Javanese sister species. More often the focus is on mobile organisms, i.e. butterflies and birds, which have endemic species on the larger and more isolated Eastern islands, e.g. Sumba, Timor, or Wetar, while the islands of Bali to Flores are usually inhabited by the same widespread species30,31,32,56. This highlights the need for more biogeographical studies on less vagile taxa, e.g. frogs, molluscs and flightless arthropods from the Lesser Sunda Islands.
Patterns of organisms transgressing Wallace’s Line are often complex, with frequent back-colonizations32,57,58. Organisms with weaker dispersal abilities may exhibit fewer transgressions leading to an increase in diversification rates59 or resulting in a single radiation60,61. The range expansion of Trigonopterus is rather unidirectional and shows a conspicuous absence of back-colonization, similar to a pattern found in another group of flightless weevils33. It is remarkable that a genus of flightless insects managed to colonize an area spanning >9,000 km from east to west, including expanses of deep open ocean of the Western Pacific. While our reconstructions show high conservatism to a given terrane, inter-island dispersal does occur and seems to increase among smaller close-by islands, i.e. the Lesser Sunda Islands. Additionally our biogeographical model shows strong support for distance limiting dispersal in this taxon. Few means of dispersal appear possible, but presumably drift by sea currents is the most likely one. Little is known on the biology of Trigonopterus, but presumably larvae develop in dead wood like many other Cryptorhynchinae62. It is thus possible that these weevils get swept to the sea with floating twigs or other substrate and are then carried as flotsam to another shore. Sea currents in the region of the Indonesian Throughflow52,63,64 are subject to seasonal and tectonic changes, but in general the South Equatorial Current follows the New Guinea coastline from East to West. During the Oligocene this would have meant a direct connection from the area of New Guinea to the Sunda Shelf65. Once Sulawesi was formed partly blocking the gateway between Pacific and Indian oceans, patterns changed and a strong southward current in the Makassar Strait became dominant. This current would constantly bring flotsam from Sulawesi to the Lesser Sunda Islands. Basic biological information would allow the modelling of dispersal by sea currents, e.g. the duration and substrate of larval development, survival in sea water etc.66; in the absence of these data we can only observe that the patterns found are well compatible with a scenario of sea current dispersal. It would also explain the unidirectionality since a drift from the Sunda Arc northwards to Sulawesi would be highly unlikely with the present system of sea currents.
The exchange between Sahul and Sunda floras is dominated by dispersal from West to East and zoochorous lineages were overrepresented among dispersers suggesting mainly birds or bats as primary means of long-distance dispersal between the two shelves67,68. However, a study on the entire flora of Java finds it closely related to Sulawesi and the Lesser Sunda Islands, which is attributed to climatic reasons by Van Welzen et al.69. This is not a suitable explanation for the similar patterns exhibited by Trigonopterus weevils usually restricted to pockets of everwet rainforest, even in highly seasonal areas. Possibly, these common patterns between plants and weevils are more strongly influenced by other factors than climate, i.e. the sea currents directed form Sulawesi to Java and the Lesser Sunda Islands.
Reconstructions of Pleistocene vegetation types70,71 are at odds with local radiations of Trigonopterus in East Java and the Lesser Sunda Islands dating back to the Miocene (Fig. 1, S1, clades H, I, J) as these areas are supposedly covered with seasonally dry forests during the Pleistocene. Even today, Trigonopterus species are highly restricted to small pockets of wet montane forests and these must have persisted to explain current patterns. Most likely, climate models are not detailed enough to account for localized areas of high precipitation on isolated mountains, but these montane pockets do matter as long-term refugia for an endemic arthropod fauna. Thus, geology, distance and sea currents were probably the dominant factors shaping Trigonopterus biogeography, not climate.
Materials and Methods
Taxon sampling and DNA sequencing
We sampled 96 Trigonopterus species from Sundaland and the Lesser Sunda Islands representing all species known from that region except for two species from Java and one from Flores43 (Supplement 3). We added 94 representatives from neighbouring areas (Sulawesi: 52; Moluccas: 13; New Guinea: 29). Altogether, our dataset contains 190 Trigonopterus species (one represented by two subspecies) plus four outgroup representatives of other cryptorhynchine species from Australia, New Guinea and Java (Critomerus iliacus (Pascoe); Microporopterus cf. setosus Voss; Ouporopterus squamiventris Lea; Miocalles sp.). All Trigonopterus species from the Sunda region and some from New Guinea possess a valid name43,72, while most of the species from Sulawesi are currently being revised and described. Undescribed species are referred to by unique species numbers that will be provided in future taxonomic treatments. All the species were monophyletic in a phylogeny using CO1 data of multiple specimens per species and also well delineated by male genital characters. The high diversity of Trigonopterus in New Guinea42 was represented by the selection of species used earlier to represent major lineages38.
Extraction, PCR, sequencing and alignment
DNA was extracted non-destructively using the DNeasy and NucleoSpin 96 Tissue kits (Qiagen, Hilden; Macherey-Nagel, Düren, Germany). Voucher specimens are used for subsequent taxonomic studies (e.g.43,72) and are stored in our museum collections (MZB, SMNK, SMNK). Primers and PCR conditions are listed in Table S10; for additional information on our standard protocols see http://zsm-entomology.de/wiki/The_Beetle_D_N_A_Lab. We sequenced 12 gene fragments with an alignment of 6,511 base pairs (bps) consisting of fragments from cytochrome oxidase subunit 1 (CO1) (2 non-overlapping fragments), 16S mtDNA, Arginine kinase (AK), carbamoyl-phosphate synthetase 2 (CAD) (3 non-overlapping fragments), Elongation factor 1α (EF1 α), Enolase (EN), Histone 4 (H4), 18S and 28S. Sequences were edited using Sequencher 4.10.1 (GeneCodes Corp., Ann Arbor, MI, USA). Coding genes were aligned in MUSCLE73, noncoding genes in MAFFT version 774. Sequences were color-coded by amino acid in MEGA675 and checked for stop codons. The final dataset was generated in Sequence Matrix 1.7.2 (Ref. 76).
Phylogenetic analyses
Relationships among Trigonopterus species were reconstructed using maximum parsimony (MP), maximum likelihood (ML) and Bayesian Inference (BI). TNT 1.177,78 was used for MP analyses with “traditional search” and 1000 replicates. ML analyses were conducted with the best partitioning scheme selected in PartitionFinder 1.1.1 (79; Table S11) using IQ-Tree80,81 with standard parameters. We conducted 1000 bootstrap replicates (BS) which are indicating the level of support at each node. A calculated BS ≥ 70 was considered to indicate strong support for a given clade in the ML analyses82. BI analyses were conducted in MrBayes 3.283, using the different partitions recovered by PartitionFinder; instead of using the selected substitution models, reversible-jump MCMC has been used to explore the entire space of substitution models84. We sampled 60 million generations of two independent runs consisting of eight Markov Chains Monte Carlo (MCMC) sampling every 5000th generation. Split-frequencies and log-likelihood curves were examined in Tracer 1.5 (http://beast.bio.ed.ac.uk/Tracer); the first 2.000 out of 12.000 trees sampled were removed as burnin and a 50% majority rule consensus tree was constructed based on the remaining trees. A calculated PP ≥ 0.95 was considered to indicate strong support for a given clade in the BI analyses85.
Estimation of divergence times
Phylogenetic trees were dated using the Bayesian relaxed clock method implemented in BEAST 1.8.186. Each of the Markov chain Monte Carlo (MCMC) analyses were run for 100 million generations to reach stationarity, with trees and model parameters sampled from the stationary posterior distribution. Stationarity was assessed using the program Tracer version 1.5. Exploratory analyses have shown a much too old dating when using published rates for Coleoptera, e.g. calibration with 0.0177 substitutions per site per Ma per lineage50 placed the earliest node of Trigonopterus at 66.05 Ma (95% HPD: 49.99–89.51 Ma) which would predate the node of non-cryptorhynchine Molytinae and the cryptorhynchine Acalles, an early genus of the group producing Trigonopterus (87; unpublished data). This is not entirely surprising as the rate of molecular evolution in flightless beetles, especially groups inhabiting stable habitats might be highly accelerated (Ikeda et al., 2012; Vogler & Timmermans, 2012; Mitterboeck & Adamowicz, 2013)93,94,95 which could in part also explain the high interspecific divergence in Trigonopterus38. There are no Trigonopterus fossils and general weevil fossils cannot be used due to the uncertain monophyly of Cryptorhynchinae and the unresolved placement of Trigonopterus within this subfamily. The only time-calibrated tree of Curculionidae available87 did not recover Cryptorhynchinae as monophyletic, perhaps because >30% of the data was missing. Furthermore, the scant fossil record of this subfamily does not offer a taxon to which Trigonopterus could be safely attributed. As a result, we were not able to use a secondary calibration for the Trigonopterus radiation. Here we used the emergence of more extensive amounts of land of New Guinea at 30 Ma11,52,54 (analysis 1, Fig. S2) and alternatively the emergence of Java at 10 Ma – following a complete inundation by shallow sea11,44 (analysis 2, Fig. 1) as two geological constraints. Although these two calibration points enforce maximum ages for these islands, they are the best source of calibrations currently available for this group. We performed two analyses with two geological calibration points (see above). The analyses were performed under a Speciation: Birth-Death model using an estimated relaxed clock rate (uncorrelated lognormal) since the hypothesis of a strict molecular clock was tested and rejected in PAUP* (p-value < 0.001)88. The MCMC parameters were fixed to 100 million generations with sampling every 10.000th generation (20.000 trees) and discarding 4.000 trees as burn-in. In order to reduce the computational time and the parameter space to explore, we fixed the best BI topology from which we removed all outgroups by manually editing the .xml file created in BEAUTi 1.8.186. A 50% majority rule consensus tree was created in TreeAnnotator 1.8.1.
Ancestral state reconstruction
We used Mesquite89 for a Maximum Parsimony approach to infer ancestral areas. This is free of assumptions on likelihood of dispersal and/or vicariance, on distance between areas and on changing historical landmass configurations. Map figures were made using GeoMapApp v3.4.190 http://www.geomapapp.org and modified in Adobe Photoshop CS2.
Biogeographical model fitting and ancestral range estimation
We used the R package BioGeoBEARS to test a variety of model-based approaches that incorporate information on clade age, distance and/or connectivity between areas45,91. BioGeoBEARS requires as inputs (1) a dated phylogeny, which we derived from BEAST and (2) a file of geographic ranges indicating the presence/absence of each species in each area in the analysis. The sampling localities were grouped into the following 12 discrete areas: Bali, Borneo, Flores, E-Java, W-Java, Lombok, Moluccas, New Guinea, Sulawesi, Sumatra, Sumbawa, Philippines (Figs 1 and 2). We allowed for a maximum of three areas at each node, resulting in a total of 298 possible states (geographic ranges) in the state space. We chose to have no more than three areas at a node because most Trigonopterus species are micro-endemics. In the analysis, a few nodes were assigned a multi-area range, e.g. the ancestor is hypothesized as having occupied a joint range of New Guinea and W-Java (in clade A of analysis 1). We find this joint range highly unlikely for the same reasons; species of Trigonopterus usually occur in very restricted areas, e.g. a single mountain range or a single island and readily speciate, even given small geographic barriers.
A map of the region was produced in GeoMapApp v3.4.1 (http://www.geomapapp.org) and the shape of each area was traced in QGIS software (www.qgis.org). Distances between the areas were defined in QGIS (Fig. 1), with the closest distance between two areas measured in kilometres. These distances were used in the constrained-distance-dependent dispersal matrices (Tables S2–S7). The resulting shape files were imported into R to construct distance and connectivity matrices between regions.
There are three basic models that we used: first, the dispersal extinction cladogenesis (DEC) model of Ree et al.92 and Ree & Smith46; second, the DEC + J model, incorporating founder event speciation45,91 where ranges can expand to novel areas at cladogenesis; third, DEC + J + x incorporating additionally distance-dependent dispersal. We implemented 12 different variants of these models with two different sets of constraints: (1) the DEC model, unconstrained, (2) DEC + J unconstrained, (3) DEC model with dispersal constrained to adjacent areas, (4) the DEC + J model with dispersal constrained to adjacent areas, (5) DEC model with dispersal constrained to adjacent areas except for the Sunda Arc (W-Java, E-Java, Bali, Lombok, Sumbawa, Flores), (6) the DEC + J model with dispersal constrained to adjacent areas except for the Sunda Arc (W-Java, E-Java, Bali, Lombok, Sumbawa, Flores), (7) DEC + x dispersal is not limited to adjacent areas (unconstrained dispersal) and modified by the exponent x describing distance, (8) DEC + J + x dispersal is not limited to adjacent areas (unconstrained dispersal), which adds jump dispersal, also modified by distance x. (9) DEC + x model, where dispersal is limited to adjacent areas, (10) DEC + J + x model, where dispersal is limited to adjacent areas, (11) DEC + x model with dispersal constrained to adjacent areas except for the Sunda Arc and (12) DEC + J + x model with dispersal constrained to adjacent areas except for the Sunda Arc (Suppl. Fig. 6).
The “+x” model in BioGeoBEARS takes the distance in kilometres from area A to B and multiplies the rates of dispersal by the exponent of the distance from A to B. For a more thorough explanation of the “+x” models in BioGeoBEARS. Here we used a value of 9,999 km to indicate an unlikely dispersal event in our distance matrix and turned off the constraint matrix in BioGeoBEARS. This configuration would allow dispersal between areas set to a distance of 9,999 km but these dispersal events would be highly unlikely. By turning off the hard constraints, it makes dispersal possible but unlikely given the long dispersal distances. We also rescaled our distance matrix to see if that had any effect on the results. We repeated the analyses with the distances re-scaled to a maximum distance 1 and the others fractions of it, achieving the same results as an input of raw distances in kilometres. We found that rescaling the distance matrix had no effect and so proceeded with using distance in kilometres.
Time-stratified analyses
Here we used the 12 models outlined above and additionally we used four separate time intervals for which we measured the distances between areas based on the geological reconstructions of Hall11. As our stem node dated to c. 14 Ma we used two longer intervals (15–10 Ma and 10–5 Ma) and two shorter, more recent intervals from 5–2.58 Ma and 2.58 Ma until present. The most recent of these time slices used the modern geological configuration. In the 15–10 Ma time slice, we excluded W-Java, E-Java, Bali, Lombok, Sumbawa and Flores. All areas were allowed in the other time slices. For the manual dispersal probability multipliers we used a value of 0.5 to reflect the uncertainty11,12,52 in the formation of land for Bali, Lombok, Sumbawa and Flores. This was only implemented during the 10–5 Ma time slice. For the distance measurements between areas we used the reconstructions of Hall11 except for the 2.58 Ma to modern time slice, where we used the modern configuration. Figures were digitized and converted into the Spherical Mercator projection to take measurements. See supporting information for constraints, distance matrixes and shape files.
Statistical model choice
The DEC-type models are nested within DEC + J type models; these models could be compared in pairwise fashion using the LRT, a chi-squared test with one degree of freedom (DEC 2 free parameters, DEC + J 3 free parameters). For comparing models on a particular dataset, Akaike information criterion (AIC) and AIC corrected for sample size (AICc) were used. AICc-based weights were calculated and used to calculate relative model probabilities as percentages (Burham & Anderson, 2002)96 indicating which model received the most support given the geographic range data.
Additional Information
Accession codes: Sequences have been deposited in EMBL Protein Knowledgebase database under accession numbers HG939627-HG940360, HE615185-HE616067, LM655408-LM656033, LN884315-LN885085, LN880278-LN880449. Data on genetic material contained in this paper is published for non-commercial use only. Utilization for purposes other than non-commercial scientific research may infringe the conditions under which the genetic resources were originally accessed and should not be undertaken without contacting the corresponding author of the paper and/or seeking permission from the original provider of the genetic material.
How to cite this article: Tänzler, R. et al. Macroevolution of hyperdiverse flightless beetles reflects the complex geological history of the Sunda Arc. Sci. Rep. 6, 18793; doi: 10.1038/srep18793 (2016).
References
Lohman, D. J., de Bruyn, M., Page, T., von Rintelen, K., Hall, R., Ng, P. K. L., Shi, H.-T., Carvalho, G. P. & von Rintelen, T. (2011). Biogeography of the Indo-Australian archipelago. Annu. Rev. Ecol. Evol. Syst. 42, 205–226, 10.1146/annurev-ecolsys-102710-145001.
Klaus, S., Selvandran, S., Goh, J. W., Wowor, D., Brandis, D., Koller, P., Schubart, C. D., Streit, B., Meier, R., Ng, P. K. L. & Yeo, D. C. J. (2013) Out of Borneo: Neogene diversification of Sundaic freshwater crabs (Crustacea: Brachyura: Gecarcinucidae: Parathelphusa). Journal of Biogeography 40, 63–74, 10.1111/j.1365-2699.2012.02771.x.
von Rintelen, T., Stelbrink, B., Marwoto, R. M. & Glaubrecht, M. (2014). A Snail Perspective on the Biogeography of Sulawesi, Indonesia: Origin and Intra-Island Dispersal of the Viviparous Freshwater Gastropod Tylomelania. PloS One 9(6), e98917.
Gonzalez, P., Su, Y. C., Siler, C. D., Barley, A. J., Sanguila, M. B., Diesmos, A. C. & Brown, R. M. (2014). Archipelago colonization by ecologically dissimilar amphibians: Evaluating the expectation of common evolutionary history of geographical diffusion in co-distributed rainforest tree frogs in islands of Southeast Asia. Molecular phylogenetics and evolution 72, 35 41.
Siler, C. D., Diesmos, A. C., Alcala, A. C. & Brown, R. M. (2011). Phylogeny of Philippine slender skinks (Scincidae: Brachymeles) reveals underestimated species diversity, complex biogeographical relationships and cryptic patterns of lineage diversification. Molecular Phylogenetics and Evolution 59(1), 53 65
Toussaint, E. F. A., Hall, R., Monaghan, M. T., Sagata, K., Ibalim, S., Shaverdo, H. V. & Balke, M. (2014). The towering orogeny of New Guinea as a trigger for arthropod megadiversity. Nature communications 5, 4001, 10.1038/ncomms5001.
Pacey, A., Macpherson, C. G. & McCaffrey, K. J. (2013). Linear volcanic segments in the central Sunda Arc, Indonesia, identified using Hough transform analysis: implications for arc lithosphere control upon volcano distribution. Earth and Planetary Science Letters 369, 24 33.
Rensch, B. (1936) Die Geschichte des Sundabogens: Eine tiergeographische Untersuchung. Berlin: Gebr. Borntraeger.
van Oosterzee, P. (1997). Where Worlds Collide: The Wallace Line. Ithaca, NY: Cornell Univ. Press. 234 pp.
Whitmore, T. C. (1981). Wallace’s Line and Plate Tectonics. Oxford, UK: Clarendon Press. 200 pp.
Hall, R. (2013). The palaeogeography of Sundaland and Wallacea since the Late Jurassic. Journal of Limnology 72(2), 1 17.
Spakman, W. & Hall, R. (2010). Surface deformation and slab–mantle interaction during Banda arc subduction rollback. Nature Geoscience 3(8), 562 566.
Rutherford, E., Burke, K. & Lytwyn, J. (2001). Tectonic history of Sumba Island, Indonesia, since the Late Cretaceous and its rapid escape into the forearc in the Miocene. Journal of Asian Earth Sciences 19(4), 453 479.
Voris, H. K. (2000). Maps of Pleistocene sea levels in Southeast Asia: shorelines, river systems and time durations. Journal of Biogeography 27, 1153 1167.
Woodruff, D. (2010). Biogeography and conservation in Southeast Asia: how 2.7 million years of repeated environmental fluctuations affect today’s patterns and the future of the remaining refugial-phase biodiversity. Biodiversity and Conservation 19, 919 941.
Mayr, E. (1944) Wallace’s line in the light of recent zoogeographic studies. Q. Rev. Biol. 19, 1 14.
Wallace, A. R. (1860) On the zoological geography of the Malay Archipelago. Zool. J. Linn. Soc. 4, 172 184.
Huxley, T. H. (1868) On the classification and distribution of the Alectoromorphae and Heteromorphae. Proc. Zool. Soc. Lond. 1868, 294 319.
Slik, J. F., Aiba, S. I., Bastian, M., Brearley, F. Q., Cannon, C. H., Eichhorn, K. A. & Zweifel, N. (2011). Soils on exposed Sunda Shelf shaped biogeographic patterns in the equatorial forests of Southeast Asia. Proceedings of the National Academy of Sciences 108(30), 12343 12347.
Gorog, A. J., Sinaga, M. H. & Engstrom, M. D. (2004). Vicariance or dispersal? Historical biogeography of three Sunda shelf murine rodents (Maxomys surifer, Leopoldamys sabanus and Maxomys whiteheadi). Biol. J. Linn. Soc. 81, 91 109.
Lohman, D. J., Peggie, D., Pierce, N. E. & Meier, R. (2008). Phylogeography and genetic diversity of a widespread Old World butterfly, Lampides boeticus (Lepidoptera: Lycaenidae). BMC evolutionary biology 8(1), 301.
Meijaard, E. (2003). Mammals of south-east Asian islands and their Late Pleistocene environments. Journal of Biogeography 30(8), 1245 1257.
Schmitt, L., Kitchener, D. & How, R. (1995). A genetic perspective of mammalian variation and evolution in the Indonesian Archipelago: biogeographic correlates in the fruit bat genus Cynopterus. Evolution 49, 399 412.
Luo, S.-J., Kim, J.-H., Johnson, W. E., van der Walt, J., Martenson, J. et al. (2004). Phylogeography and genetic ancestry of tigers (Panthera tigris). PLoS Biol. 2, 2275 2293.
Esselstyn, J. A., Timm, R. M. & Brown, R. M. (2009). Do geological or climatic processes drive speciation in dynamic archipelagos? The tempo and mode of diversification in Southeast Asian shrews. Evolution 63(10), 2595 2610.
Clouse, R. M. & Giribet, G. (2010). When Thailand was an island–the phylogeny and biogeography of mite harvestmen (Opiliones, Cyphophthalmi, Stylocellidae) in Southeast Asia. Journal of Biogeography 37(6), 1114 1130.
Clouse, R. M., Janda, M., Blanchard, B., Sharma, P., Hoffmann, B. D., Andersen, A. N. & Wheeler, W. C. (2014). Molecular phylogeny of Indo‐Pacific carpenter ants (Hymenoptera: Formicidae, Camponotus) reveals waves of dispersal and colonization from diverse source areas. Cladistics.
Condamine, F. L., Toussaint, E. F., Cotton, A. M., Genson, G. S., Sperling, F. A. & Kergoat, G. J. (2013) Fine-scale biogeographical and temporal diversification processes of peacock swallowtails (Papilio subgenus Achillides) in the Indo-Australian Archipelago. Cladistics 29(1), 88 111.
Condamine, F. L., Toussaint, E. F. A., Clamens, A.-L., Genson, G., Sperling, F. A. H. & Kergoat, G. J. (2015) Deciphering the evolution of birdwing butterflies 150 years after Alfred Russel Wallace. Scientific Reports 5, 11860.
Müller, C. J. & Beheregaray, L. B. (2010). Palaeo island-affinities revisited–Biogeography and systematics of the Indo-Pacific genus Cethosia Fabricius (Lepidoptera: Nymphalidae). Molecular Phylogenetics and Evolution 57(1), 314 326.
Müller, C. J., Wahlberg, N. & Beheregaray, L. B. (2010). ‘After Africa’: the evolutionary history and systematics of the genus Charaxes Ochsenheimer (Lepidoptera: Nymphalidae) in the Indo-Pacific region. Biological Journal of the Linnean Society 100(2), 457 481.
Müller, C. J., Matos-Maraví, P. F. & Beheregaray, L. B. (2013). Delving into Delias Hübner (Lepidoptera: Pieridae): fine-scale biogeography, phylogenetics and systematics of the world’s largest butterfly genus. Journal of Biogeography 40(5), 881 893.
Toussaint, E. F. A., Tänzler, R., Rahmadi, C., Balke, M. & Riedel, A. (2015). Biogeography of Australasian flightless weevils (Curculionidae, Celeuthetini) suggests permeability of Lydekker’s and Wallace’s Lines. Zoologica Scripta. Early Online, 10.1111/zsc.12127.
Wahlberg, N. & Rubinoff, D. (2011). Vagility across Vanessa (Lepidoptera: Nymphalidae): mobility in butterfly species does not inhibit the formation and persistence of isolated sister taxa. Systematic Entomology 36(2), 362 370.
O’Grady, P. & DeSalle, R. (2008). Out of Hawaii: the origin and biogeography of the genus Scaptomyza (Diptera: Drosophilidae). Biology Letters 4(2), 195 199.
Bellemain, E. & Ricklefs, R. E. (2008). Are islands the end of the colonization road? Trends in Ecology & Evolution 23(8), 461 468.
Evans, B. J., Brown, R. M., McGuire, J. A., Supriatna, J., Andayani, N., Diesmos, A. & Cannatella, D. C. (2003). Phylogenetics of fanged frogs: testing biogeographical hypotheses at the interface of the Asian and Australian faunal zones. Systematic Biology 52(6), 794 819.
Tänzler, R., Toussaint, E. F. A., Suhardjono, Y. R., Balke, M. & Riedel, A. (2014). Multiple transgressions of Wallace’s Line explain diversity of flightless Trigonopterus weevils on Bali. Proceedings of the Royal Society B 281, 20132528.
Köhler, F. & Criscione, F. (2013). Plio-Pleistocene out-of-Australia dispersal in a camaenid land snail. Journal of Biogeography 4010, 1971–1982.
Riedel, A. (2010). One of a thousand - a new species of Trigonopterus (Coleoptera, Curculionidae, Cryptorhynchinae) from New Guinea. Zootaxa 2403, 59 68.
Riedel, A., Daawia, D. & Balke, M. (2010) Deep cox1 divergence and hyperdiversity of Trigonopterus weevils in a New Guinea mountain range (Coleoptera, Curculionidae). Zoologica Scripta 39, 63 74.
Tänzler, R., Sagata, K., Surbakti, S., Balke, M. & Riedel, A. (2012) DNA barcoding for community ecology - how to tackle a hyperdiverse, mostly undescribed Melanesian fauna. PLoS One 7, e28832, 10.1371/journal.pone.0028832.
Riedel, A., Tänzler, R., Balke, M., Rahmadi, C. & Suhardjono, Y. R. (2014) Ninety-eight new species of Trigonopterus weevils from Sundaland and the Lesser Sunda Islands. ZooKeys 467, 1–162, 10.3897/zookeys.467.8206.
Hall, R. (2012). Sundaland and Wallacea: geology, plate tectonics and palaeogeography. In Biotic evolution and environmental change in Southeast Asia (eds Gower, D. J., Johnson, K. G., Richardson, J. E., Rosen, B. R., Rüber, L. & Williams, S. T. ) pp. 32–78, Cambridge: Cambridge University Press.
Matzke, N. J. (2014). Model selection in historical biogeography reveals that founder-event speciation is a crucial process in island clades. Systematic Biology, syu056.
Ree, R. H. & Smith, S. A. (2008). Maximum-likelihood inference of geographic range evolution by dispersal, local extinction and cladogenesis. Syst. Biol. 57, 4 14.
Andújar, C., Serrano, J. & Gómez-Zurita, J. (2012). Winding up the molecular clock in the genus Carabus (Coleoptera: Carabidae): assessment of methodological decisions on rate and node age estimation. BMC evolutionary biology 12(1), 40.
Bauzà-Ribot, M. M., Juan, C., Nardi, F., Oromí, P., Pons, J. & Jaume, D. (2012). Mitogenomic phylogenetic analysis supports continental-scale vicariance in subterranean thalassoid crustaceans. Current Biology 22(21), 2069 2074.
Brower, A. V. Z. (1994). Rapid morphological radiation and convergence among races of the butterfly Heliconius erato inferred from mitochondrial DNA evolution. Proceedings of the National Academy of Sciences USA 91, 6491 6495.
Papadopoulou, A., Anastasiou, I. & Vogler, A. P. (2010) Revisiting the Insect Mitochondrial Molecular Clock: The Mid-Aegean Trench Calibration. Molecular Biology and Evolution 27(7), 1659 1672.
Stelbrink, B., Albrecht, C., Hall, R. & von Rintelen, T. (2012) The biogeography of Sulawesi revisited: is there evidence for a vicariant origin of taxa on Wallace’s ‘anomalous island’? Evolution 66, 2252–2271, 10.1111/j.1558-5646.2012.01588.x.
Hall, R. (2009). Southeast Asia’s changing palaeogeography. Blumea 54(1–3), 148 161.
Barberi, F., Bigioggero, B., Boriani, A., Cattaneo, M., Cavallin, A., Cioni, R., Eva, C., Gelmini, R., Giorgetti, F., Iaccarino, S., Innocenti, F., Marinelli, G., Slejko, D. & Sudradjat, A. (1987) The island of Sumbawa: A major structural discontinuity in the Indonesian Arc. Boll. Soc. Geol. It. 106, 547 620.
van Ufford, A. Q. & Cloos, M. (2005) Cenozoic tectonics of New Guinea. Am. Assoc. Petr. Geol. B 89, 119 140.
Nishimura, S., Harjono, H. & Suparka, S. (1992). The Krakatau Islands: the geotectonic setting. GeoJournal 28(2), 87 98.
Outlaw, D. C. & Voelker, G. (2008). Pliocene climatic change in insular Southeast Asia as an engine of diversification in Ficedula flycatchers. Journal of Biogeography 35(4), 739–752, 10.1111/j.1365-2699.2007.01821.x.
Jønsson, K. A., Irestedt, M., Fuchs, J., Ericson, P. G., Christidis, L., Bowie, R. C. & Fjeldså, J. (2008) Explosive avian radiations and multi-directional dispersal across Wallacea: evidence from the Campephagidae and other Crown Corvida (Aves). Molecular phylogenetics and evolution 47(1), 221–236, 10.1016/j.ympev.2008.01.017.
Balke, M., Ribera, I., Hendrich, L., Miller, M. A., Sagata, K., Posman, A. & Meier, R. (2009). New Guinea highland origin of a widespread arthropod supertramp. Proceedings of the Royal Society B: Biological Sciences 276(1666), 2359 2367.
Bacon, C. D., Michonneau, F., Henderson, A. J., McKenna, M. J., Milroy, A. M. & Simmons, M. P. (2013). Geographic and taxonomic disparities in species diversity: dispersal and diversification rates across Wallace’s line. Evolution 67(7), 2058–2071, 10.1111/evo.12084.
Brown, G. K., Nelson, G. & Ladiges, P. Y. (2006). Historical biogeography of Rhododendron section Vireya and the Malesian Archipelago. Journal of Biogeography 33(11), 1929–1944, 10.1111/j.1365-2699.2006.01548.x.
Webb, C. O. & Ree, R. (2012). Historical biogeography inference in Malesia. In Biotic evolution and environmental change in Southeast Asia (eds Gower, D. J., Johnson, K. G., Richardson, J. E., Rosen, B. R., Rüber, L. & Williams, S. T. ) pp. 138–163, Cambridge: Cambridge University Press.
Lyal, C. H. C. (2014). 3.7.7 Molytinae Schoenherr, 1823. In: Leschen, R. A. B. & Beutel, R. G. (Eds) Handbook of Zoology, Coleoptera, Beetles Volume 3: Morphology and Systematics (Phytophaga). DeGruyter, Berlin, pp. 529 570.
Gordon, A. L. & Fine, R. A. (1996) Pathways of water between the Pacific and Indian oceans in the Indonesian seas. Nature 379, 146–149, 10.1038/379146a0.
Gordon, A. L. (2005). The Indonesian Seas. Oceanography of the Indonesian sea and their throughflow. Oceanography 18(4), 14 27.
Kuhnt, W., Holbourn, A., Hall, R., Zuvela, M. & Käse, R. (2004). Neogene history of the Indonesian throughflow. In: Clift, P., Kuhnt, W., Wang, P. & Hayes, D. (Eds), Continent-Ocean Interactions within East Asian Marginal Seas. American Geophysical Union, Washington, DC, pp. 299 320.
Treml, E. A., Roberts, J. J., Chao, Y., Halpin, P. N., Possingham, H. P. & Riginos, C. (2012). Reproductive output and duration of the pelagic larval stage determine seascape-wide connectivity of marine populations. Integrative and Comparative Biology 52(4), 525 537.
Crayn, D. M., Costion, C. & Harrington, M. G. (2015). The Sahul–Sunda floristic exchange: dated molecular phylogenies document Cenozoic intercontinental dispersal dynamics. Journal of Biogeography 42(1), 11–24.
Richardson, J. E., Costion, C. M. & Müllner, A. N. (2012). The Malesian floristic interchange: plant migration patterns across Wallace′s Line. In Biotic evolution and environmental change in Southeast Asia (eds Gower, D. J., Johnson, K. G., Richardson, J. E., Rosen, B. R., Rüber, L. & Williams, S. T. ) pp. 138–163, Cambridge: Cambridge University Press.
Van Welzen, P. C., Parnell, J. A. & Slik, J. F. (2011). Wallace’s Line and plant distributions: two or three phytogeographical areas and where to group Java? Biological Journal of the Linnean Society 103(3), 531 545.
de Bruyn, M., Stelbrink, B., Morley, R. J., Hall, R., Carvalho, G. R., Cannon, C. H. & Von Rintelen, T. (2014). Borneo and Indochina are major evolutionary hotspots for Southeast Asian biodiversity. Systematic Biology 63(6), 879 901.
Cannon, C. H., Morley, R. J. & Bush, A. B. G. (2009). The current refugial rainforests of Sundaland are unrepresentative of their biogeographic past and highly vulnerable to disturbance. Proceedings of the National Academy of Sciences USA 106, 11188 11193.
Riedel, A., Sagata, K., Surbakti, S., Tänzler, R. & Balke, M. (2013) One hundred and one new species of Trigonopterus weevils from New Guinea. ZooKeys 280, 1–150, 10.3897/zookeys.280.3906.
Edgar, R. C. (2004). MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC bioinformatics 5(1), 113.
Katoh, K. & Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular biology and evolution 30(4), 772 780.
Tamura, K., Stecher, G., Peterson, D., Filipski, A. & Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular biology and evolution 30(12), 2725–2729.
Vaidya, G., Lohman, D. J. & Meier, R. 2011. SequenceMatrix: concatenation software for the fast assembly of multi‐gene datasets with character set and codon information. Cladistics 27(2), 171 180.
Goloboff, P. A., Farris, J. S. & Nixon, K. C. (2007) Tree analysis using new technology. Version 1.1.
Goloboff, P. A., Farris, J. S. & Nixon, K. C. (2008) TNT, a free program for phylogenetic analysis. Cladistics 24, 774 786.
Lanfear, R., Calcott, B., Ho, S. Y. & Guindon, S. (2012). PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Molecular biology and evolution 29(6), 1695–1701.
Minh, B. Q., Nguyen, M. A. T. & von Haeseler, A. (2013) Ultrafast approximation for phylogenetic bootstrap. Molecular biology and evolution 30, 1188–1195, 10.1093/molbev/mst024.
Nguyen, L. T., Schmidt, A. H., von Haeseler, A. & Minh, B. Q. (2015) IQ-TREE: A fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Molecular Biology and Evolution 32, 268–274, 10.1093/molbev/msu300.
Hillis, D. M. & Bull, J. J. (1993) An Empirical Test of Bootstrapping as a Method for Assessing Confidence in Phylogenetic Analysis. Systematic Biology 42(2), 182–192, 10.1093/sysbio/42.2.182.
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Höhna, S., Larget, B., Liu, L., Suchard, M. A. & Huelsenbeck, J. P. (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61(3), 539 542.
Huelsenbeck, J. P., Larget, B. & Alfaro, M. E. (2004) Bayesian phylogenetic model selection using reversible jump Markov chain Monte Carlo. Molecular Biology and Evolution 21(6), 1123 1133.
Erixon, P., Svennblad, B., Britton, T. & Oxelman, B. (2003) Reliability of Bayesian Posterior Probabilities and Bootstrap Frequencies in Phylogenetics. Systematic Biology 52(5), 665–673, 10.1080/10635150390235485.
Drummond, A. J., Suchard, M. A., Xie, D. & Rambaut, A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29, 1969 1973.
McKenna, D. D., Sequeira, A. S., Marvaldi, A. E. & Farrell, B. D. (2009) Temporal lags and overlap in the diversification of weevils and flowering plants. PNAS 106, 7083–7088, 10.1073/pnas.0810618106.
Swofford, D. L. 2002 PAUP*. Phylogenetic Analysis using Parsimony (*and other methods), version 4.0b10 [Computer software and manual]. Sunderland, MA: Sinauer Associates.
Maddison, W. & Maddison, D. (2009). Mesquite: a modular system for evolutionary analysis. Version 2.6. Computer program and documentation distributed by the author.
Ryan, W. B. F., Carbotte, S. M., Coplan, J. O., O’Hara, S., Melkonian, A., Arko, R., Weissel, R. A., Ferrini, V., Goodwillie, A., Nitsche, F., Bonczkowski, J. & Zemsky, R. (2009) Global Multi-Resolution Topography synthesis. Geochemistry, Geophysics, Geosystems 10, Q03014, 10.1029/2008GC002332.
Matzke, N. J. (2013) Probabilistic historical biogeography: new models for founder-event speciation, imperfect detection and fossils allow improved accuracy and model-testing. Diss. University of California, Berkeley, 2013.
Ree, R. H., Moore, B. R., Webb, C. O. & Donoghue, M. J. (2005). A likelihood framework for inferring the evolution of geographic range on phylogenetic trees. Evolution 59, 2299 2311.
Ikeda, H., Nishikawa, M. & Sota, T. 2012 Loss of flight promotes beetle diversification. Nat. Commun. 3, 648. (10.1038/ncomms1659).
Vogler, A. P., & Timmermans, M. J. 2012 Speciation: Don’t Fly and Diversify? Curr. Biol. 22, R284-R286. (http://dx.doi.org/10.1016/j.cub.2012.03.015).
Mitterboeck, T. F. & Adamowicz, S. J. 2013 Flight loss linked to faster molecular evolution in insects. Proc. R. Soc. Lond. B 280. (10.1098/rspb.2013.1128).
Burnham, K. P., & Anderson, D. R. (2002). Model selection and multimodel inference: a practical information-theoretic approach. Springer Science & Business Media. 488 pp.
Acknowledgements
We thank LIPI (Indonesian Institute of Sciences), RISTEK (Ministry of State for Research and Technology, Indonesia) and the Indonesian Department of Forestry for providing relevant permits. Voucher specimens will be deposited in LIPI Division of Zoology Collection, formerly the Museum Zoologicum Bogoriense (Cibinong, Indonesia) and the Museum of Natural History Karlsruhe (Germany). We thank two anonymous reviewers for valuable comments. Last, but not least, the field work would not have been possible without the generous hospitality and help of many local people and we thank all of them very warmly. This work was funded by the German Research Foundation, DFG (RI 1817/1-1, 3-1, 3-3). M.H.V.D. was funded through NSF award DBI #1402102.
Author information
Authors and Affiliations
Contributions
A.R., M.B. and M.H.V.D. conceived the ideas; A.R. and Y.R.S. collected the specimens; R.T. carried out the molecular work and figure design; R.T. and E.F.A.T. did the phylogenetic analyses, M.H.V.D. designed and performed the BioGeoBEARS analyses and statistical model choice and R.T. performed the parsimony biogeographical analysis and lineage accumulation plot; All authors made significant comments on and improvements to the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Tänzler, R., Van Dam, M., Toussaint, E. et al. Macroevolution of hyperdiverse flightless beetles reflects the complex geological history of the Sunda Arc. Sci Rep 6, 18793 (2016). https://doi.org/10.1038/srep18793
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep18793
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.