Abstract
Link prediction is a fundamental problem with applications in many fields ranging from biology to computer science. In the literature, most effort has been devoted to estimate the likelihood of the existence of a link between two nodes, based on observed links and nodes’ attributes in a network. In this paper, we apply several representative link prediction methods to reconstruct the network, namely to add the missing links with high likelihood of existence back to the network. We find that all these existing methods fail to identify the links connecting different communities, resulting in a poor reproduction of the topological and dynamical properties of the true network. To solve this problem, we propose a community-based link prediction method. We find that our method has high prediction accuracy and is very effective in reconstructing the inter-community links.
Similar content being viewed by others
Introduction
Many complex systems can be naturally described by complex networks, which has largely deepened our understanding of the structure of real systems. For example, many topological properties, such as small-world1, scale-free2, assortativity3, community4 and rich club5, have been uncovered in not only the social and technology systems we are using everyday6,7,8,9,10,11, but also the biology systems within our bodies12,13,14. In addition, network representation is useful from practical point of view. It allows us to optimize the systems for higher functionality15,16,17 and predict the future evolution of real systems18,19. Link prediction is one of these significant research problems20. It aims to estimate the likelihood of the existence of a link between two nodes, based on observed links and nodes’ attributes in a network. With this problem solved, a large amount of cost in lab experiment for identifying the missing data could be reduced20.
Link prediction methods assume that similar nodes are those that have similar connectivity patterns. Therefore, the essential problem in link prediction is to objectively estimate the similarity between nodes21. Up to now, many similarity metrics on link prediction have been proposed. The most straightforward method is the so-called Common Neighbor index which directly computes the number of overlapped neighbors between two nodes to determine their similarity22. This index, though simple, has many shortcomings. It is strongly biased to the large degree nodes and it works poorly in sparse networks. To solve these problems, many other methods, such as Jaccard23, Resource Allocation24, Local Path methods25 etc, are designed. Recently, some attention has also been paid to study link prediction in weighted26,27, directed28,29, bipartite30,31 networks. Moreover, some link prediction methods have been introduced to detect the spurious connections in complex networks32.
In order to quantify the quality of link prediction, the index called area under the receiver operating characteristic curve (AUC) is usually used33. In practice, it calculates the probability that a true link has a higher link prediction score than a nonexisting link. In the case of predicting missing links, the predicted links need to be added to the observed networks to obtain the reconstructed networks20. The  index can only reflect the fraction of corrected links added to the network, but cannot capture whether the reconstructed network has the same or similar structural and dynamical properties as the true network. This is especially important in the networks with community structure34. It can happen in such networks that a link prediction method correctly identifies many missing links, but completely neglects those links connecting different communities. These inter-community links actually play an important role in the networks. They characterize the interactions between different clusters35. They are also strongly related to many global network properties such as average shortest path and the betweenness centrality36. Without these links, some dynamical properties such as bond percolation will be largely distorted37.
index can only reflect the fraction of corrected links added to the network, but cannot capture whether the reconstructed network has the same or similar structural and dynamical properties as the true network. This is especially important in the networks with community structure34. It can happen in such networks that a link prediction method correctly identifies many missing links, but completely neglects those links connecting different communities. These inter-community links actually play an important role in the networks. They characterize the interactions between different clusters35. They are also strongly related to many global network properties such as average shortest path and the betweenness centrality36. Without these links, some dynamical properties such as bond percolation will be largely distorted37.
In this paper, we apply several representative link prediction methods to reconstruct complex networks, namely to add the missing links with high likelihood of existence back to the networks. Even though large  is achieved, the reconstructed networks from these existing methods are found to be very different from the true networks, especially in terms of the average betweenness of the predicted links. This result indicates that the missing inter-community links are seldom captured by the existing link prediction methods. To solve this problem, we propose a community-based link prediction method. Our method can effectively identify the inter-community links by slightly sacrificing the prediction accuracy. The final obtained network can thus well reproduce the structural and dynamical properties of the true network.
is achieved, the reconstructed networks from these existing methods are found to be very different from the true networks, especially in terms of the average betweenness of the predicted links. This result indicates that the missing inter-community links are seldom captured by the existing link prediction methods. To solve this problem, we propose a community-based link prediction method. Our method can effectively identify the inter-community links by slightly sacrificing the prediction accuracy. The final obtained network can thus well reproduce the structural and dynamical properties of the true network.
Results
We consider an undirected network  where
where  is the set of nodes and
is the set of nodes and  is the set of links. In link prediction, the original links
is the set of links. In link prediction, the original links  are first randomly divided into two parts: the training set (
are first randomly divided into two parts: the training set ( ) and the probe set (
) and the probe set ( ). The training set contains
). The training set contains  of the original links and the link prediction methods run on it. The probe set consists of the remaining
of the original links and the link prediction methods run on it. The probe set consists of the remaining  of the original links (The results of other division ratios are shown in SI). The probe set is used to test the accuracy of the link prediction methods. The accuracy is usually measured by the
of the original links (The results of other division ratios are shown in SI). The probe set is used to test the accuracy of the link prediction methods. The accuracy is usually measured by the  value (see the Methods section for details), the higher the better. Besides accuracy, we consider also whether the link prediction methods can effectively recover the structural properties of the original network. Normally, the link prediction methods predict missing links by assigning each unconnected node pair a score which estimates the likelihood for each node pair to have a missing link between them. An accurate link prediction method will assign high score to the true missing links and low score to the nonexistent links. Unfortunately, for most of the existing link prediction methods, there is no obvious score gap between the true missing links and nonexistent links. Therefore, in order to reconstruct the network, one has to assume that the number of true missing links
value (see the Methods section for details), the higher the better. Besides accuracy, we consider also whether the link prediction methods can effectively recover the structural properties of the original network. Normally, the link prediction methods predict missing links by assigning each unconnected node pair a score which estimates the likelihood for each node pair to have a missing link between them. An accurate link prediction method will assign high score to the true missing links and low score to the nonexistent links. Unfortunately, for most of the existing link prediction methods, there is no obvious score gap between the true missing links and nonexistent links. Therefore, in order to reconstruct the network, one has to assume that the number of true missing links  is roughly known. In this fashion, one can add
is roughly known. In this fashion, one can add  top-ranking links in the link prediction methods to the observed network to reconstruct the predicted network. The approach is widely used in the literature38,39. Consistent with the previous works, we also assume that we know roughly the total number of true missing links. The
top-ranking links in the link prediction methods to the observed network to reconstruct the predicted network. The approach is widely used in the literature38,39. Consistent with the previous works, we also assume that we know roughly the total number of true missing links. The  node pairs (L = |EP|) with the highest score (denoted as the “predicted links”) will be added to the training set
node pairs (L = |EP|) with the highest score (denoted as the “predicted links”) will be added to the training set  to obtain the reconstructed network G′(V, E′). A well-performed link prediction method should not only aim at achieving a high
to obtain the reconstructed network G′(V, E′). A well-performed link prediction method should not only aim at achieving a high  value, but also make the structural properties of G′(V, E′) close to G(V, E).
value, but also make the structural properties of G′(V, E′) close to G(V, E).
In this paper, we focus on the networks with community structure. According to the definition, the nodes within a community are densely connected while the nodes across communities are much more sparsely connected. In this kind of networks, the inter-community links are in general more difficult to be predicted. Without these inter-community links, the average shortest path length of the reconstructed networks would be much higher than the original networks and the transportation dynamics40 in this network would be much slower and congested in the reconstructed networks. In order to solve this problem, we propose a community-based link prediction method. We first detect the communities by using the  algorithm41 in the training set. Then the similarity scores between unconnected node pairs are computed by some classic local similarity measures (i.e. the CN or RA methods, see the Methods section for definitions). We also consider three global link prediction methods32,39,42, the results are similar to those of CN and RA (see Supplementary Information (SI)). A tunable parameter β ∈[0, 1] is proposed to combine the information of communities and node similarity for link prediction. In practice, the node pairs are classified as intra-community pairs and inter-community pairs. Within each classification, the node pairs are ranked in descending order according to the similarity measures.
algorithm41 in the training set. Then the similarity scores between unconnected node pairs are computed by some classic local similarity measures (i.e. the CN or RA methods, see the Methods section for definitions). We also consider three global link prediction methods32,39,42, the results are similar to those of CN and RA (see Supplementary Information (SI)). A tunable parameter β ∈[0, 1] is proposed to combine the information of communities and node similarity for link prediction. In practice, the node pairs are classified as intra-community pairs and inter-community pairs. Within each classification, the node pairs are ranked in descending order according to the similarity measures.  controls the probability that the intra-community node pairs ranked higher than the inter-community node pairs (see the Methods section for details). This method is inspired by ref. 43 but used here for a different goal. For convenience, when the method is combined with common neighbor similarity, it is called community-based CN method (CBCN). Similarly, it is called community-based RA method (CBRA) when it is combined with the resource allocation similarity. The illustration of the method is shown in Fig. 1. Like previous works43, we adopt
controls the probability that the intra-community node pairs ranked higher than the inter-community node pairs (see the Methods section for details). This method is inspired by ref. 43 but used here for a different goal. For convenience, when the method is combined with common neighbor similarity, it is called community-based CN method (CBCN). Similarly, it is called community-based RA method (CBRA) when it is combined with the resource allocation similarity. The illustration of the method is shown in Fig. 1. Like previous works43, we adopt  to evaluate the accuracy of the link prediction. In addition, we propose to monitor the average edge-betweenness
to evaluate the accuracy of the link prediction. In addition, we propose to monitor the average edge-betweenness  of the predicted links (calculated by adding those predicted links to the network). If the average edge-betweenness is high, more inter-community links are predicted (For the solid evidences, see SI). In fact, measuring the average betweenness of the reconstructed network is also a good evaluation metric for this issue. Despite some quantitative difference, the results are qualitatively consistent with the results when
of the predicted links (calculated by adding those predicted links to the network). If the average edge-betweenness is high, more inter-community links are predicted (For the solid evidences, see SI). In fact, measuring the average betweenness of the reconstructed network is also a good evaluation metric for this issue. Despite some quantitative difference, the results are qualitatively consistent with the results when  is used (see results in SI).
is used (see results in SI).
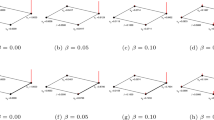
The illustration of the community-based link prediction method.
The network on the left is the network consisting of the links in the training set. The nodes within one community are marked by the same color. The solid links represent the observed links and the dashed links stand for the predicted links. When β = 0, the inter-community missing links are ranked higher than the intra-community missing links in the prediction list. Therefore, mainly inter-community links are added to the network by the link prediction method. When β = 1, the intra-community missing links are ranked higher than the inter-community missing links in the prediction list and mainly intra-community links are added to the network. When β = 0.05, the results are mixed, both inter- and intra-community missing links are added to the network. The similarity measure used in this toy network is CN.
We first test our method in a classical artificial network: GN-benchmark network35 which is widely used in the research of community structure. In the GN-benchmark network, n = 128 nodes equally distribute in 4 communities and each node has on average  links where
links where  is the average number of neighbors within the same community (
is the average number of neighbors within the same community ( ) and
) and  is the average number of neighbors between different communities (
is the average number of neighbors between different communities ( ). As
). As  increases, the community structure of network becomes clear. Given an observed network, the obtained similarity score between nodes is deterministic if CN and RA similarity measurements are applied. However, the community detection algorithm has randomness. Therefore, there is some stochasticity in the link prediction process coming from the community detection algorithm. In this paper, we use the extremal optimization (EO) algorithm to detect communities. As stated in ref. 41, the performance of this algorithm is rather stable. Therefore, the stochasticity of the link prediction process is expected to be relatively small. We perform several times of realizations and find that the variance is much smaller than the mean value. Therefore, we mainly report the results of the mean value of different realizations.
increases, the community structure of network becomes clear. Given an observed network, the obtained similarity score between nodes is deterministic if CN and RA similarity measurements are applied. However, the community detection algorithm has randomness. Therefore, there is some stochasticity in the link prediction process coming from the community detection algorithm. In this paper, we use the extremal optimization (EO) algorithm to detect communities. As stated in ref. 41, the performance of this algorithm is rather stable. Therefore, the stochasticity of the link prediction process is expected to be relatively small. We perform several times of realizations and find that the variance is much smaller than the mean value. Therefore, we mainly report the results of the mean value of different realizations.
In Fig. 2, we show the dependence of  and
and  on
on  under different
under different  . The CBCN and CBRA are used in Fig. 2(a–d), respectively. One can see that
. The CBCN and CBRA are used in Fig. 2(a–d), respectively. One can see that  increases with
increases with  , indicating that the links within the communities are easier to be predicted. The results of CBCN and CBRA are similar and the increment of
, indicating that the links within the communities are easier to be predicted. The results of CBCN and CBRA are similar and the increment of  is more significant when the community structure is more obvious (i.e. larger
is more significant when the community structure is more obvious (i.e. larger  ). This result is consistent with a recent finding in ref. 43. In Fig. 2(a,b), the dashed lines mark the
). This result is consistent with a recent finding in ref. 43. In Fig. 2(a,b), the dashed lines mark the  of the original CN and RA methods (without
of the original CN and RA methods (without  to adjust the ranking of the intra- and inter-community missing links). One can see that the
to adjust the ranking of the intra- and inter-community missing links). One can see that the  of CBCN and CBRA can be respectively higher than the
of CBCN and CBRA can be respectively higher than the  of CN and RA when
of CN and RA when  is large.
is large.
The influence of β on AUC and 〈B〉 in the GN-benchmark networks.
(a–d) are the results of CBCN and CBRA, respectively. The solid lines are the results of the community-based link prediction methods (CBCN and CBRA) and the dashed lines are the results of the classic link prediction methods (CN and RA). The results are averaged over 100 independent realizations.
In Fig. 2(c,d), it shows that  actually decreases with
actually decreases with  . This is natural as a larger
. This is natural as a larger  means more intra-community missing links are ranked higher, thus the predicted links are mainly within communities. In Fig. 2(c,d) the dashed lines mark the
means more intra-community missing links are ranked higher, thus the predicted links are mainly within communities. In Fig. 2(c,d) the dashed lines mark the  of the links in the probe set. Clearly, if one only considers
of the links in the probe set. Clearly, if one only considers  , β = 1 is the optimal solution. However, this setting of
, β = 1 is the optimal solution. However, this setting of  would make
would make  of the predicted links smaller than that of the true missing links. A good link prediction method should not only have high
of the predicted links smaller than that of the true missing links. A good link prediction method should not only have high  but also make
but also make  of the predicted links close to that of the true missing links. Interestingly, we observe that when
of the predicted links close to that of the true missing links. Interestingly, we observe that when  is large, a small change in
is large, a small change in  can result in a significant decrease in
can result in a significant decrease in  but little influence on
but little influence on  . This observation indicates the possibility to adjust
. This observation indicates the possibility to adjust  for a satisfactory results in both
for a satisfactory results in both  and
and  .
.
We also examine our method on four real networks: ZK is a social network in the zahcary karate club44, NS is the largest connected component of a co-authorship network of scientists who are publishing on the topic of network science45, Email is an email network of an university built by regarding each email address as a node and linking two nodes if there is an email communication between them46, C.elegans is a neural network of the worm Caenorhadities elegans with each neuron as a node and each synapse or gap junction as a link47. All of these real networks are widely used in the literature and the basic structural properties of them are listed in Table 1. Here we use them to examine our methods. Figure 3 shows the performance of the community-based link prediction methods on these real networks. One can see that the results are qualitatively the same as those in the GN-benchmark networks. In these real networks, as the community structure is not as obvious as the GN-benchmark, the effect of  on
on  is even smaller, especially after β > 0.1. However, the influence of
is even smaller, especially after β > 0.1. However, the influence of  on
on  is still strong.
is still strong.
The influence of β on AUC and 〈B〉 in four real networks.
(a–d) are the results of CBCN and CBRA, respectively. The solid lines are the results of the community-based link prediction methods (CBCN and CBRA) and the dashed lines are the results of the classic link prediction methods (CN and RA). The results are averaged over  independent realizations.
independent realizations.
We denote  as the
as the  that can make
that can make  of the predicted links the same as that of the true missing links (i.e. the links in the probe set). Accordingly, the
of the predicted links the same as that of the true missing links (i.e. the links in the probe set). Accordingly, the  under
under  is denoted as
is denoted as  . The quantitative results of
. The quantitative results of  and
and  in four real networks are reported in Table 1. Clearly, the
in four real networks are reported in Table 1. Clearly, the  of CBCN and CBRA can still be higher than the
of CBCN and CBRA can still be higher than the  of CN and RA, respectively.
of CN and RA, respectively.
To further understand the performance of each method, we compute the number of correctly predicted inter- and intra-links and the number of inter- and intra-links in the predicted links (results are shown in SI). We find that when the existing link prediction methods are used in GN-benchmark, the number of inter-links in the predicted links is almost zero, indicating that these existing methods tend to neglect inter-links. On the contrary, CBCN and CBRA have many inter-links in the predicted links. However, if we look at the number of correctly predicted inter-links in our methods, the number is also small. This is because the inter-links are sparsely and randomly connected in GN-benchmark (i.e. almost form no triangle) and it is difficult for CBCN and CBRA to capture their similarity to other links. In real networks, however, the inter-links form more triangles than thus are easier to be predicted. We test the NS real network with clear community structure (collaboration network between network scientists). We find that CN and RA can correctly predict 17.6 and 30.0 inter-links while CBCN and CNRA can correctly predict 23.7 and 31.5 inter-links (For more detailed results in NS network, see SI). These results indicates that CBCN and CNRA can respectively outperforms CN and RA in real networks as well.
In Fig. 4, we further investigate the influence of  on
on  and
and  in the GN-benchmark networks. In Fig. 4(a,b), one can see that
in the GN-benchmark networks. In Fig. 4(a,b), one can see that  has an abrupt change after kin > 10. After this value,
has an abrupt change after kin > 10. After this value,  significantly increases with
significantly increases with  . This is because when the community structure is obvious (kin > 10), we don’t have to sacrifice too much
. This is because when the community structure is obvious (kin > 10), we don’t have to sacrifice too much  and a large
and a large  can already make
can already make  close to the true value. In Fig. 4(c,d), we show the dependence of
close to the true value. In Fig. 4(c,d), we show the dependence of  on
on  . One can see that when
. One can see that when  is large,
is large,  is very close to the
is very close to the  of the original CN or RA. However, when
of the original CN or RA. However, when  is relatively small,
is relatively small,  can be much smaller than
can be much smaller than  of CN or RA. This is because when
of CN or RA. This is because when  is small,
is small,  needs to be adjusted to a very small value in order to keep
needs to be adjusted to a very small value in order to keep  of the predicted links the same as the real links (as shown in Fig. 2). In this case, a large amount of
of the predicted links the same as the real links (as shown in Fig. 2). In this case, a large amount of  needs to be sacrificed for a higher
needs to be sacrificed for a higher  .
.
The influence of κin on β* and AUC* in the GN-benchmark networks.
(a–d) are the results of CBCN and CBRA, respectively. The solid lines are the results of the community-based link prediction methods (CBCN and CBRA) and the dashed lines are the results of the classic link prediction methods (CN and RA). The results are averaged over 100 independent realizations.
So far, we have already shown that adjusting  in the community-based link prediction methods can indeed help the methods predict more high-betweenness links in the networks. A natural question to ask at this point is how to choose
in the community-based link prediction methods can indeed help the methods predict more high-betweenness links in the networks. A natural question to ask at this point is how to choose  in real use. Even though
in real use. Even though  can be chosen at the value where
can be chosen at the value where  of the predicted links becomes the same as the real links. However, as
of the predicted links becomes the same as the real links. However, as  of the real links is unknown information, the above strategy seems to be an inapplicable way. To solve this problem, one has to learn the optimal
of the real links is unknown information, the above strategy seems to be an inapplicable way. To solve this problem, one has to learn the optimal  from the observed data. To mimic this process, we use a so-called threefold validation where a small part (usually
from the observed data. To mimic this process, we use a so-called threefold validation where a small part (usually  of all links) is moved from the previously introduced training set
of all links) is moved from the previously introduced training set  to a learning set
to a learning set  48. The threefold validation is usually used to avoid model over-fitting in machine learning. In our case, by checking at which
48. The threefold validation is usually used to avoid model over-fitting in machine learning. In our case, by checking at which  the predicted links from
the predicted links from  can have the same
can have the same  as the links in
as the links in  , one can determine the estimated optimal parameter
, one can determine the estimated optimal parameter  .
.
One concern for the learning process is that the missing links may largely change the structural properties. To check this, we first conduct the community detection algorithm (EO algorithm) on the original true network and denote the obtained communities as the “true detected communities”. Then we randomly remove a fraction of links from the true network to obtain the observed network. We do again the community detection algorithm on the observed network and compute the fraction of nodes classified correctly by comparing the obtained communities with the so-called “true detected communities”. We find that the fraction of nodes classified correctly is rather high, especially when the community structure is obvious (correct rate is over 80% when kin ≥ 10). Moreover, we compare  with
with  determined with
determined with  in Fig. 4(a,b). One can see that
in Fig. 4(a,b). One can see that  at different
at different  .
.
The learned optimal parameter  is then used to predict missing links based on
is then used to predict missing links based on  which are then compared with entries in
which are then compared with entries in  to finally measure the link prediction accuracy
to finally measure the link prediction accuracy  . The results are shown in Fig. 4(c,d). One can see that
. The results are shown in Fig. 4(c,d). One can see that  is indeed close to
is indeed close to  . As discussed above, the
. As discussed above, the  is usually too small when kin < 13, which directly results in a low
is usually too small when kin < 13, which directly results in a low  in link prediction. Therefore, we propose an additional constraint in the learning process: when determining the optimal
in link prediction. Therefore, we propose an additional constraint in the learning process: when determining the optimal  with the learning set
with the learning set  , we also monitor the prediction
, we also monitor the prediction  of these links in
of these links in  (denoted as
(denoted as  ). In order to make sure the optimal
). In order to make sure the optimal  will not be too small, we assume that at most we can sacrifice
will not be too small, we assume that at most we can sacrifice  of the accuracy. Here, we define the
of the accuracy. Here, we define the  of the original method CN or RA as
of the original method CN or RA as  . If before
. If before  drops to
drops to  of
of  , the predicted links can have the same
, the predicted links can have the same  as the links in
as the links in  ,
,  is chosen as this crossover point. If not,
is chosen as this crossover point. If not,  is chosen as the value where
is chosen as the value where  equals to
equals to  of
of  . The
. The  obtained in this way is denoted as “constrained
obtained in this way is denoted as “constrained  ”. The results of the constrained
”. The results of the constrained  and its prediction accuracy “constrained
and its prediction accuracy “constrained  ” are shown in Fig. 4 as well. So far, we have discussed three parameters:
” are shown in Fig. 4 as well. So far, we have discussed three parameters:  ,
,  and constrained
and constrained  . A summary of these three parameters is given in Table 2. Note that even though the amount of missing links is not known, the estimation of
. A summary of these three parameters is given in Table 2. Note that even though the amount of missing links is not known, the estimation of  and constrained
and constrained  will not be influenced. This is because
will not be influenced. This is because  and constrained
and constrained  are obtained from the learning process in which the amount of links in the learning set
are obtained from the learning process in which the amount of links in the learning set  is known.
is known.
 and Constrained
and Constrained  .
.Moreover, we study whether the structural and dynamical properties of the reconstructed networks from CBCN and CBRA are truly closer to the true networks. We take into account six indices, including the average shortest path of the networks  , clustering coefficient (
, clustering coefficient ( )47, assortativity coefficient (
)47, assortativity coefficient ( )3, congestibility (
)3, congestibility ( )49, synchronizability (
)49, synchronizability ( )50 and spreading ability (
)50 and spreading ability ( )51. The results of different link prediction methods are listed in Table 3. The original real networks are denoted as
)51. The results of different link prediction methods are listed in Table 3. The original real networks are denoted as  . We first randomly divide the links in
. We first randomly divide the links in  to three parts: training set
to three parts: training set  (with 80% of the links), learning set
(with 80% of the links), learning set  (with 10% of the links) and probe set
(with 10% of the links) and probe set  (with 10% of the links). We apply the community-based link prediction methods to compute the constrained
(with 10% of the links). We apply the community-based link prediction methods to compute the constrained  with
with  and
and  . Then we do
. Then we do  to obtain a complete
to obtain a complete  . We apply the community-based link prediction methods with the constrained
. We apply the community-based link prediction methods with the constrained  on the complete
on the complete  . The
. The  number of links with the highest link prediction score are then added to
number of links with the highest link prediction score are then added to  to create the reconstructed network
to create the reconstructed network  . We also create the reconstructed networks with
. We also create the reconstructed networks with  arbitrarily set as
arbitrarily set as  and
and  and denote these networks as
and denote these networks as  and
and  , respectively. For comparison, the reconstructed networks with the traditional link prediction methods (e.g. CN and RA) are denoted as
, respectively. For comparison, the reconstructed networks with the traditional link prediction methods (e.g. CN and RA) are denoted as  . From Table 3, we can see that the reconstructed networks from the community-based link prediction methods (i.e.
. From Table 3, we can see that the reconstructed networks from the community-based link prediction methods (i.e.  ,
,  and
and  ) have more similar network properties to the real network
) have more similar network properties to the real network  than those obtained by the traditional link prediction methods (
than those obtained by the traditional link prediction methods ( ). The best results sometimes appear in
). The best results sometimes appear in  and
and  . However, when
. However, when  is closest to
is closest to  ,
,  is very different from
is very different from  and vice versa.
and vice versa.  keeps a reasonable trade-off between these two methods:
keeps a reasonable trade-off between these two methods:  best reproduces the network properties of
best reproduces the network properties of  in many cases; when
in many cases; when  is not the best,
is not the best,  is the closest one to the best. These results confirm the importance of the parameter learning process.
is the closest one to the best. These results confirm the importance of the parameter learning process.
Finally, we discuss the computational complexity of our method. The method is actually a combination of local link prediction algorithm and the community detection algorithm. For the local link prediction algorithm such as CN and RA, the computational complexity is  where
where  is the number of nodes and
is the number of nodes and  is the mean degree of the network. In this paper, we use the extremal optimization (EO) algorithm for community detection, with computational complex
is the mean degree of the network. In this paper, we use the extremal optimization (EO) algorithm for community detection, with computational complex  . Apparently, the computational complexity in our method is mainly determined by the community detection algorithm. If the method is applied to large networks, one can choose a faster community detection algorithm, such as the method in ref. 52 with complexity
. Apparently, the computational complexity in our method is mainly determined by the community detection algorithm. If the method is applied to large networks, one can choose a faster community detection algorithm, such as the method in ref. 52 with complexity  in which
in which  is the number of edges in the network.
is the number of edges in the network.
Discussion
Predicting the missing or future links is a very important research topic itself and has applications in many different domains. Although many link prediction methods have been proposed in the literature, they consider all the missing links homogeneous (i.e. all the missing links are considered equally important). In this paper, we argue that in the networks with community structure, the links connecting different communities are actually of more significance and more difficult to be predicted. We propose a community-based link prediction method which allows us to predict more missing inter-community links (with high edge-betweenness) in both artificial and real networks. The results show that our method can predict more high betweenness links without losing much link prediction accuracy. As the community-based link prediction method has a parameter to tune, we propose a learning process to determine the optimal parameter. We finally apply the community-based link prediction method to reconstruct networks. The results show that the reconstructed networks by our method have very similar network properties with the real networks.
Even though our paper tries to solve a specific problem, it points out several long-neglected important issues in link prediction research: (i) Links in the network are not with equal importance. The algorithms should give priority to those important links. (ii) Prediction results should be evaluated not only by accuracy but also by how much the predicted links can recover the properties of the true network. (iii) The parameters in the link prediction algorithms should be estimated via a learning process before applied to real prediction. These issues will encourage researchers to reconsider the existing works in link prediction and may inspire a series of more effective algorithms in the future.
In this paper, we proposes an effective method to predict the inter-community links. Compared to the existing methods which all fail to predict the inter-community links (especially when the community structure is obvious), our method has a large proportion of inter-community links in the top ranking. We admit that the improved precision of these inter-community links is not high, this is because those links have a very low probability of existing. However, by including more inter-community links in the prediction list, we manage to obtain reconstructed networks with closer topological properties to the true networks. Predicting important links in networks is a scientific problem which cannot be completely solved in one paper, it surely asks for more studies in the future. Therefore, our paper raises up some important questions for future research. The method in this paper use the classic EO community algorithm to detect communities. An interesting question would be comparing the performance of different community algorithms in helping link prediction algorithms identify inter-community links. In the networks without clear community structure, the links with high edge-betweenness are still more important than the low edge-betweenness links. In these networks, the method proposed in this paper cannot be directly applied as it relies on the community detection method. Therefore, how to predict high edge-betweenness links in networks without community structure is an important extension. Finally, our study highlights the fact that the missing links are not with equal importance. Besides betweenness, the importance of links can be measured by other properties such as degree-product, clustering coefficient, link salience53 etc. We hope the method in this paper will shed some light on designing methods to predict these kinds of important links in complex networks.
Methods
Classic link prediction algorithms
We use two representative classic link prediction algorithms in this paper: common neighbors (CN) and resource allocation (RA). After the network data is divided into the training set  and probe set
and probe set  , these two methods generate the predicted links by estimating the similarity values between different node pairs in
, these two methods generate the predicted links by estimating the similarity values between different node pairs in  . We denote the set of neighbors of node
. We denote the set of neighbors of node  by
by  .
.
CN simply measures the similarity between node  and node
and node  with the number of overlapped neighbors,
with the number of overlapped neighbors,

RA is a variant of CN. In RA, the weight of each common neighbor is negatively proportional to its degree. The similarity is thus computed as

where  is the degree of node
is the degree of node  and
and  is the set of the common neighbors between
is the set of the common neighbors between  and
and  . After obtaining
. After obtaining  for each node pairs, the missing links is ranked by sorting
for each node pairs, the missing links is ranked by sorting  in descending order.
in descending order.
Community detection
The community detection method in the paper is the EO method41. It detects communities by optimizing the modularity  with a heuristic search. The modularity
with a heuristic search. The modularity  is defined as
is defined as

where  is the contribution of individual node
is the contribution of individual node  given a certain partition into communities.
given a certain partition into communities.  is the number of links node
is the number of links node  has with nodes in the same community
has with nodes in the same community  ,
,  is the community which node
is the community which node  belongs to.
belongs to.  is the degree of node
is the degree of node  and
and  is the fraction of links that have one or two nodes inside of the community
is the fraction of links that have one or two nodes inside of the community  .
.  is the number of the links in the network.
is the number of the links in the network.
Community-based link prediction method
After computing  , the node pairs are classified into two sets according to the community detection results: intra-community node pairs and inter-community node pairs. The node pairs in each set are ranked according to
, the node pairs are classified into two sets according to the community detection results: intra-community node pairs and inter-community node pairs. The node pairs in each set are ranked according to  in descending order. The ranking list in intra-community node pairs is denoted as Rinter and the ranking list in inter-community node pairs is denoted as Rinter. The parameter
in descending order. The ranking list in intra-community node pairs is denoted as Rinter and the ranking list in inter-community node pairs is denoted as Rinter. The parameter  is used when Rinter and Rinter are combined. Initially,
is used when Rinter and Rinter are combined. Initially,  is empty. The node pairs are then moved from Rinter and Rinter to
is empty. The node pairs are then moved from Rinter and Rinter to  one by one from top to bottom. In each step, Rinter is picked with probability
one by one from top to bottom. In each step, Rinter is picked with probability  and
and  is picked with probability
is picked with probability  . For instance, if there is already
. For instance, if there is already  node pairs in
node pairs in  and in next step Rinter is picked, highest ranked node pair in Rinter is removed and placed in the
and in next step Rinter is picked, highest ranked node pair in Rinter is removed and placed in the  position in
position in  . Note that the ranking list Rinter and Rinter become shorter and shorter while the ranking list
. Note that the ranking list Rinter and Rinter become shorter and shorter while the ranking list  becomes longer and longer. The procedure is terminated if both Rinter and Rinter are empty.
becomes longer and longer. The procedure is terminated if both Rinter and Rinter are empty.
Result evaluation
The results of the link prediction are evaluated by  and
and  .
.  (area under the
(area under the  curve) is a way to quantify the accuracy of prediction algorithms54. At each time, we randomly select a nonexisting link in the original network and a link in the probe set to compare their positions in
curve) is a way to quantify the accuracy of prediction algorithms54. At each time, we randomly select a nonexisting link in the original network and a link in the probe set to compare their positions in  . After n times of comparison, there are n′ times the probe set links have a higher rank and n″ times the probe set links have the same rank as the nonexisting links, then the
. After n times of comparison, there are n′ times the probe set links have a higher rank and n″ times the probe set links have the same rank as the nonexisting links, then the  value is
value is

Besides  , we considered another important metric called Precision. It is defined as the fraction of correctly predicted links in the top-
, we considered another important metric called Precision. It is defined as the fraction of correctly predicted links in the top- ranking list. Here,
ranking list. Here,  is set as the total number of missing links. The results are shown in SI. Despite some quantitative difference, the results of precision are qualitatively consistent with that of
is set as the total number of missing links. The results are shown in SI. Despite some quantitative difference, the results of precision are qualitatively consistent with that of  (i.e. prediction accuracy increases with
(i.e. prediction accuracy increases with  ).
).
 is defined as the average betweenness of the predicted links when they are added to the networks. The predicted links are just
is defined as the average betweenness of the predicted links when they are added to the networks. The predicted links are just  number of top ranking links in
number of top ranking links in  . The betweenness of a link
. The betweenness of a link  is defined as the ratio of the shortest paths which pass through the edge
is defined as the ratio of the shortest paths which pass through the edge  among all the shortest paths in the network,
among all the shortest paths in the network,

 is the number of shortest routes between node
is the number of shortest routes between node  and
and  ,
,  is the number of the shortest paths between node
is the number of the shortest paths between node  and
and  which pass through the edge
which pass through the edge  .
.
Additional Information
How to cite this article: Zhang, P. et al. The reconstruction of complex networks with community structure. Sci. Rep. 5, 17287; doi: 10.1038/srep17287 (2015).
References
Milgram, S. The small world problem. Psychol. Today 2, 60–67 (1967).
Barabási, A. L. & Albert, R. Emergence of scaling in random networks. Science 286, 509–512 (1999).
Newman, M. E. Assortative mixing in networks. Phys. Rev. Lett. 89, 208701 (2002).
Radicchi, F. et al. Defining and identifying communities in networks. Proc. Natl. Acad. Sci. USA 101, 2658–2663 (2004).
Zhou, S. & Mondragón, R. J. The rich-club phenomenon in the Internet topology. IEEE Commun. Lett. 8, 180–182 (2004).
Amaral, L. A. N., Scala, A., Barthélémy, M. et al. Classes of small-world networks. Proc. Natl. Acad. Sci. USA 97, 11149–11152 (2000).
Borgatti, S. P. et al. Network analysis in the social sciences. Science 323, 892–895 (2009).
Zhao, K. et al. Social network dynamics of face-to-face interactions. Phys. Rev. E 83, 056105 (2011).
Barabási, A. L., Albert, R. & Jeong, H. Scale-free characteristics of random networks: the topological of the world wide web. Physica A 281, 68–77 (2000).
Pastor-Satorras, R., Vázquez, A. & Vespignani, A. Dynamical and correlation properties of the Internet. Phys. Rev. E 87, 258701 (2001).
Barthélemy, M. Spatial networks. Phys. Rep. 499, 1–101 (2011).
Barabási, A. L. & Oltvai, Z. N. Network biology: understanding the cell’s functional organization. Nat. Rev. Genet. 5, 101–113 (2004).
Barabási, A. L., Gulbahce, N. & Loscalzo, J. Network medicine: a network-based approach to human disease. Nat. Rev. Genet. 12, 56–68 (2011).
Vidal, M., Cusick, M. E. & Barabási, A. L. Interactome networks and human disease. Cell 144, 986–995 (2011).
Newman, M. E. The structure and function of complex networks. Siam. Rev. 45, 167 (2003).
Buldyrev, S. V. et al. Networks formed from failures in interdependent networks. Nature 464, 1025–1028 (2010).
Gao, J. et al. Networks formed from interdependent networks. Nat. Phys. 8, 40–48 (2012).
Albert, R. & Barabási, A. L. Statistics mechanics of complex networks. Rev. Mod. Phys 74, 47 (2002).
Dorogovtsev, S. N. & Mendes, J. F. Evolution of networks. Adv. Phys. 51, 1079 (2002).
Lü, L. & Zhou, T. Link prediction in complex networks: A survey. Physica A 390, 1150–1170 (2011).
Lin, D. An information-theoretic definition of similarity. in Proceedings of the 15th International Conference on Machine Learning, 296–304 (Madison, Wisconsin, USA, 1998).
Lorrain, F. & White, H. C. Structural equivalence of individuals in social networks. J. Math. Sociol. 27, 49–80 (1971).
Jaccard, P. Étude comparative de la distribution florale dans une portion des Alpes et des Jura. Bull. Soc. vaudoise sci. nat. 37, 547–579 (1901).
Zhou, T., Lü, L. & Zhang, Y. C. Predicting missing links via local information. Eur. Phys. J. B. 71, 623 (2009).
Liu, W. & Lü, L. Link prediction based on local random walk. Europhys. Lett. 89, 58007 (2010).
Murata, T. & Moriyasu, S. Link prediction of social networks based on weighted proximity measures. In Proceedings of the IEEE/WIC/ACM International Conference on Web Intelligence, 85–88 (Washington, DC, USA, 2007).
Wind, D. K. & Morup, M. Link prediction in weighted networks. In Proceedings of the IEEE international workshop on machine learning for signal processing 1–6 (Stander, Spain, 2012).
Brzozowski, M. J. & Romero, D. M. Who should I follow? Recommending people in directed social networks. In Proceedings of the 5th international conference on weblogs and social media 458–461 (Barcelona, Catalonia, Spain, 2011).
Núria, R. et al. Predicting future conflict between team-members with parameter-free models of social networks. Sci. Rep. 3, 1999 ( 2013 ).
Kunegis, J., De, Luca, E. W. & Albayrak, S. The link prediction problem in bipartite networks. Computational intelligence for knowledge-based systems design 6178, 380–389 (2010).
Guimerà, R. et al. Predicting human preferences using the block structure of complex social networks. PLoS One 7, e44620 (2012).
Guimerà, R. & Sales-Pardo, M. Missing and spurious interactions and the reconstruction of complex networks. Proc. Natl. Acad. Sci. USA 106, 22073 (2009).
Hanely, J. A. & McNeil, B. J. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology 143, 29–36 (1982).
Santo, F. Community detection in graphs. Phys. Rep. 486, 75–174 (2010).
Girvan, M. & Newman, M. E. Community structure in social and biological networks. Proc. Natl. Acad. Sci. USA 99, 7821–7826 (2002).
Freeman, L. C. A set of measures of centrlity based on betweenness. Sociometry 40, 35–41 (1977).
Wu, C. et al. Multiple hybrid phase transition: Bootstrap percolation on complex networks with communities. Europhys. Lett. 107, 48001 (2014).
Stetter, O., Battaglia, D., Soriano, J. & Geisel, T. Model-free reconstruction of excitatory neuronal connectivity from calcium imaging signals. PLoS computational biology 8(8), e1002653 (2012).
Lü, L. et al. Toward link predictability of complex networks. Proc. Natl. Acad. Sci. USA 112, 2325–2330 (2015).
Zheng, J. F., Gao, Z. Y. & Zhao, X. M. Properties of transportation dynamics on scale-free networks. Physica A 373, 837–844 (2007).
Duch, J. & Arenas, A. Community detection in complex networks using extremal optimization. Phys. Rev. E 72, 027104 (2005).
Katz, L. A new status index derived from sociometric analysis. Psychometrika 18, 39 (1953).
Yan, B. & Gregory, S. Finding missing edges in networks based on their community structure. Phy. Rev. E 85, 056112 (2012).
Zachary, W. W. An Information flow model for conict and fission in small groups. J. Anthropol. Res 33, 452C473 (1977).
Newman, M. E. Finding community structure in networks using the eigenvectors of matrices. Phy. Rev. E 74, 036104 (2006).
Guimerà, R., Danon, L., Díaz-Guilera, A., Giralt, F. & Arenas, A. Self-similar community structure in a network of human interactions. Phys. Rev. E 68, 065103 (2003).
Watts, D. J. & Strogatz, S. H. Collective dynamics of ‘small-word’ networks. Nature 393, 440–442 (1998).
Feng, C. X. J., Yu, Z. G. S., Kingi, U. & Baig, M. P. Threefold vs. fivefold cross validation in one-hidden-layer and two-hidden-layer predictive neural network modeling of machining surface roughness data. J. Manuf. Syst. 24, 93–107 (2005).
Guimerà, R. et al. Optimal network topologies for local search with congestion. Phys. Rev. Lett. 89, 248701 (2002).
Arenas, A., Díaz-Guilera, A., Kurths, J., Moreno, Y. & Zhou, C. Synchronization in complex networks. Phys. Rep. 469, 93C153 (2008).
Boguá, M. & Pastor-Satorras, R. Epidemic spreading in correlated complex networks. Phy. Rev. E 66, 047104 (2002).
Wu, F. & Huberman, A. finding communities in linear time: a physics approach, Eur. Phys. J. B 38, 331 (2004).
Grady, D., Thiemann, C. & Brockmann, D. Robust classification of salient links in complex networks. Nat. Commun. 3, 864 (2012).
Fawcett, T. An introduction to ROC analysis. Pattern. Recogn. Lett. 27, 861 (2006).
Acknowledgements
This work was supported by The National Natural Science Foundation of China (Grant No. 61403037). AZ acknowledges the support from the Youth Scholars Program of Beijing Normal University (grant no. 2014NT38).
Author information
Authors and Affiliations
Contributions
P.Z., A.Z. and J.X. designed the research and wrote the manuscript. F.W. and X.W. performed the simulation. All authors analyzed the results and wrote the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Zhang, P., Wang, F., Wang, X. et al. The reconstruction of complex networks with community structure. Sci Rep 5, 17287 (2015). https://doi.org/10.1038/srep17287
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep17287
This article is cited by
-
Predicting missing links in complex networks based on common neighbors and distance
Scientific Reports (2016)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.