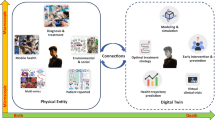
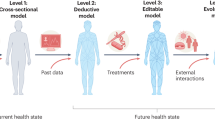
Abstract
Medical digital twins, which are potentially vital for personalized medicine, have become a recent focus in medical research. Here we present an overview of the state of the art in medical digital twin development, especially in oncology and cardiology, where it is most advanced. We discuss major challenges, such as data integration and privacy, and provide an outlook on future advancements. Emphasizing the importance of this technology in healthcare, we highlight the potential for substantial improvements in patient-specific treatments and diagnostics.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$99.00 per year
only $8.25 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
The Precision Medicine Initiative Cohort Program—Building a Research Foundation for 21st Century Medicine (NIH, 2015); https://acd.od.nih.gov/documents/reports/PMI_WG_report_2015-09-17-Final.pdf
Eddy, D. M. & Schlessinger, L. Archimedes: a trial-validated model of diabetes. Diabetes Care 26, 3093–3101 (2003).
Tomczak, K., Czerwińska, P. & Wiznerowicz, M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp. Oncol. Pozn. Pol. 19, A68–A77 (2015).
Regev, A. et al. The Human Cell Atlas. eLife 6, e27041 (2017).
Alber, M. et al. Integrating machine learning and multiscale modeling—perspectives, challenges, and opportunities in the biological, biomedical and behavioral sciences. NPJ Digit. Med. 2, 115 (2019).
Karniadakis, G. E. et al. Physics-informed machine learning. Nat. Rev. Phys. 3, 422–440 (2021).
Opportunities and Challenges for Digital Twins in Biomedical Research (National Academies of Science, Engineering and Medicine, 2023); https://nap.nationalacademies.org/catalog/26922/opportunities-and-challenges-for-digital-twins-in-biomedical-research-proceedings
Opportunities and Challenges for Digital Twins in Atmospheric and Climate Sciences: Proceedings of a Workshop—In Brief 26921 (National Academies Press, 2023); https://doi.org/10.17226/26921
Foundational Research Gaps and Future Directions for Digital Twins (National Academies of Sciences, Engineering and Medicine, 2023); https://doi.org/10.17226/26894
Wright, L. & Davidson, S. How to tell the difference between a model and a digital twin. Adv. Model. Simul. Eng. Sci. 7, 13 (2020).
Vogelsang, A. & Borg, M. Requirements engineering for machine learning: perspectives from data scientists. In Proc. 2019 IEEE 27th International Requirements Engineering Conference Workshops (REW) (eds Damian, D., Perini, A. & Lee, S.-W.) 245–251 (IEEE, 2019); https://doi.org/10.1109/REW.2019.00050
Cobelli, C. & Kovatchev, B. Developing the UVA/Padova type 1 diabetes simulator: modeling, validation, refinements and utility. J. Diabetes Sci. Technol. https://doi.org/10.1177/19322968231195081 (2023).
Breton, M. D. et al. A randomized trial of closed-loop control in children with Type 1 diabetes. N. Engl. J. Med. 383, 836–845 (2020).
Quintairos, A., Pilcher, D. & Salluh, J. I. F. ICU scoring systems. Intensive Care Med. 49, 223–225 (2023).
Dang, J. et al. Developing DELPHI expert consensus rules for a digital twin model of acute stroke care in the neuro critical care unit. BMC Neurol. 23, 161 (2023).
Lal, A. et al. Development and verification of a digital twin patient model to predict specific treatment response during the first 24 hours of sepsis. Crit. Care Explor. 2, e0249 (2020).
Cockrell, C. & An, G. Sepsis reconsidered: identifying novel metrics for behavioral landscape characterization with a high-performance computing implementation of an agent-based model. J. Theor. Biol. 430, 157–168 (2017).
Larie, D., An, G. & Cockrell, R. C. The use of artificial neural networks to forecast the behavior of agent-based models of pathophysiology: an example utilizing an agent-based model of sepsis. Front. Physiol. 12, 716434 (2021).
Ribeiro, H. A. et al. Multi-scale mechanistic modelling of the host defence in invasive aspergillosis reveals leucocyte activation and iron acquisition as drivers of infection outcome. J. R. Soc. Interface 19, 20210806 (2022).
Chasseloup, E., Hooker, A. C. & Karlsson, M. O. Generation and application of avatars in pharmacometric modelling. J. Pharmacokinet. Pharmacodyn. 50, 411–423 (2023).
Moingeon, P., Chenel, M., Rousseau, C., Voisin, E. & Guedj, M. Virtual patients, digital twins and causal disease models: paving the ground for in silico clinical trials. Drug Discov. Today 28, 103605 (2023).
Allen, R., Rieger, T. & Musante, C. Efficient generation and selection of virtual populations in quantitative systems pharmacology models. CPT Pharmacomet. Syst. Pharmacol. 5, 140–146 (2016).
Hernandez-Boussard, T. et al. Digital twins for predictive oncology will be a paradigm shift for precision cancer care. Nat. Med. 27, 2065–2066 (2021).
Stahlberg, E. A. et al. Exploring approaches for predictive cancer patient digital twins: opportunities for collaboration and innovation. Front. Digit. Health 4, 1007784 (2022).
Wu, C. et al. Integrating mechanism-based modeling with biomedical imaging to build practical digital twins for clinical oncology. Biophys. Rev. 3, 021304 (2022).
Jarrett, A. M. et al. Optimal control theory for personalized therapeutic regimens in oncology: background, history, challenges and opportunities. J. Clin. Med. 9, 1314 (2020).
Baldock, A. L. et al. From patient-specific mathematical neuro-oncology to precision medicine. Front. Oncol. 3, 62 (2013).
Jackson, P. R., Juliano, J., Hawkins-Daarud, A., Rockne, R. C. & Swanson, K. R. Patient-specific mathematical neuro-oncology: using a simple proliferation and invasion tumor model to inform clinical practice. Bull. Math. Biol. 77, 846–856 (2015).
Hawkins-Daarud, A. et al. In silico analysis suggests differential response to bevacizumab and radiation combination therapy in newly diagnosed glioblastoma. J. R. Soc. Interface 12, 20150388 (2015).
Arevalo, H. J. et al. Arrhythmia risk stratification of patients after myocardial infarction using personalized heart models. Nat. Commun. 7, 11437 (2016).
Sung, E. et al. Fat infiltration in the infarcted heart as a paradigm for ventricular arrhythmias. Nat. Cardiovasc. Res. 1, 933–945 (2022).
Cartoski, M. J. et al. Computational identification of ventricular arrhythmia risk in pediatric myocarditis. Pediatr. Cardiol. 40, 857–864 (2019).
Shade, J. K. et al. Ventricular arrhythmia risk prediction in repaired tetralogy of Fallot using personalized computational cardiac models. Heart Rhythm 17, 408–414 (2020).
O’Hara, R. P. et al. Personalized computational heart models with T1-mapped fibrotic remodeling predict sudden death risk in patients with hypertrophic cardiomyopathy. eLife 11, e73325 (2022).
Shade, J. K. et al. Predicting risk of sudden cardiac death in patients with cardiac sarcoidosis using multimodality imaging and personalized heart modeling in a multivariable classifier. Sci. Adv. 7, eabi8020 (2021).
Zhang, Y. et al. Predicting ventricular tachycardia circuits in patients with arrhythmogenic right ventricular cardiomyopathy using genotype-specific heart digital twins. eLife 10.7554/eLife.88865.2 (2023).
Ashikaga, H. et al. Feasibility of image-based simulation to estimate ablation target in human ventricular arrhythmia. Heart Rhythm 10, 1109–1116 (2013).
Prakosa, A. et al. Personalized virtual-heart technology for guiding the ablation of infarct-related ventricular tachycardia. Nat. Biomed. Eng. 2, 732–740 (2018).
Sung, E. et al. Personalized digital-heart technology for ventricular tachycardia ablation targeting in hearts with infiltrating adiposity. Circ. Arrhythm. Electrophysiol. 13, e008912 (2020).
McDowell, K. S. et al. Virtual electrophysiological study of atrial fibrillation in fibrotic remodeling. PLoS ONE 10, e0117110 (2015).
Roney, C. H. et al. In silico comparison of left atrial ablation techniques that target the anatomical, structural and electrical substrates of atrial fibrillation. Front. Physiol. 11, 1145 (2020).
Zahid, S. et al. Patient-derived models link re-entrant driver localization in atrial fibrillation to fibrosis spatial pattern. Cardiovasc. Res. 110, 443–454 (2016).
Loewe, A. et al. Patient-specific identification of atrial flutter vulnerability—a computational approach to reveal latent reentry pathways. Front. Physiol. 9, 1910 (2019).
Roney, C. H. et al. Predicting atrial fibrillation recurrence by combining population data and virtual cohorts of patient-specific left atrial models. Circ. Arrhythm. Electrophysiol. 15, e010253 (2022).
Boyle, P. M. et al. Computationally guided personalized targeted ablation of persistent atrial fibrillation. Nat. Biomed. Eng. 3, 870–879 (2019).
Ali, R. L. et al. Arrhythmogenic propensity of the fibrotic substrate after atrial fibrillation ablation: a longitudinal study using magnetic resonance imaging-based atrial models. Cardiovasc. Res. 115, 1757–1765 (2019).
Shade, J. K. et al. Preprocedure application of machine learning and mechanistic simulations predicts likelihood of paroxysmal atrial fibrillation recurrence following pulmonary vein isolation. Circ. Arrhythm. Electrophysiol. 13, e008213 (2020).
EDITH: European Virtual Human Twin (Virtual Physiological Human Institute); https://www.edith-csa.eu/
Viceconti, M. & Hunter, P. The virtual physiological human: ten years after. Annu. Rev. Biomed. Eng. 18, 103–123 (2016).
Swedish Digital Twin Consortium (SDTC); https://www.sdtc.se/
Björnsson, B. et al. Digital twins to personalize medicine. Genome Med. 12, 4 (2019).
Laubenbacher, R., Sluka, J. P. & Glazier, J. A. Using digital twins in viral infection. Science 371, 1105–1106 (2021).
Laubenbacher, R. et al. Building digital twins of the human immune system: toward a roadmap. NPJ Digit. Med. 5, 64 (2022).
Forum on Precision Immunology: Immune Digital Twins (UF Laboratory for Systems Medicine); https://systemsmedicine.pulmonary.medicine.ufl.edu/working-groups/forum-on-precision-immunology-immune-digital-twins/
Building Immune Digital Twins (Institut Pascal); https://www.institut-pascal.universite-paris-saclay.fr/en/scientific-programs/building-immune-digital-twins
EDITH CSA Deliverable 3.2: First Draft of the VHT Roadmap (EDITH Consortium, 2023); https://doi.org/10.5281/ZENODO.8200955
Gartner 2018 Hype Cycle for IT in GCC Identifies Six Technologies That Will Reach Mainstream Adoption in Five to 10 Years (Gartner, 2018); https://www.gartner.com/en/newsroom/press-releases/2018-12-13-gartner-2018-hype-cycle-for-it-in-gcc-identifies-six-technologies-that-will-reach-mainstream-adoption-in-five-to-10-years
Fitzgerald, J., Larsen, P. G., Margaria, T., Woodcock, J. & Gomes, C. in Leveraging Applications of Formal Methods, Verification and Validation, Lecture Notes in Computer Science (eds Margaria, T. & Steffen, B.) 13704 (Springer, 2022); https://doi.org/10.1007/978-3-031-19762-8
Assessing the Credibility of Computational Modeling and Simulation in Medical Device Submissions; Draft Guidance for Industry and Food and Drug Administration Staff (FDA, 2021); https://www.fda.gov/media/154985/download
Nuwer, R. US agency seeks to phase out animal testing. Nature https://doi.org/10.1038/d41586-022-03569-9 (2022).
Assessing Credibility of Computational Modeling through Verification and Validation: Application to Medical Devices (The American Society of Mechanical Engineers, 2018).
Ahmed, K. B. R., Pathmanathan, P., Kabadi, S. V., Drgon, T. & Morrison, T. M. Editorial on the FDA report on ‘Successes and opportunities in modeling & simulation for FDA’. Ann. Biomed. Eng. 51, 6–9 (2023).
Acknowledgements
R.L. acknowledges support from the US Army (ACC- APG- RTP W911NF), the National Institutes of Health (NIH) 1 R01 HL169974-01), the US Department of Defense DARPA (HR00112220038), NIH 1 R011 AI135128-01 and NIH 1 R01 HL169974-01. B.M. acknowledges support from NIH 1 R01 HL169974-01, NIH 1 R011 AI135128-01 and NIH 1 R01 HL169974-01. N.T. acknowledges support from NIH R01HL166759 and R01HL142496. I.S. acknowledges support from NIH NCI R01CA270210.
Author information
Authors and Affiliations
Contributions
The authors are listed in alphabetical order in the author list. R.L. conceived the Perspective and drafted the first outline. N.T. wrote the section on digital twins in cardiology. B.M. and I.S. contributed to the other sections. All authors reviewed and edited the final version.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Computational Science thanks A. M. Alaa and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Fernando Chirigati, in collaboration with the Nature Computational Science team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Laubenbacher, R., Mehrad, B., Shmulevich, I. et al. Digital twins in medicine. Nat Comput Sci 4, 184–191 (2024). https://doi.org/10.1038/s43588-024-00607-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s43588-024-00607-6