Abstract
It has been reported that long non-coding RNA PANDA was disregulated in varieties types of tumor, but its expression level and biological role in hepatocellular carcinoma (HCC) remains contradictory. We detected PANDA expression in two independent cohorts (48 HCC patients following liver transplantation and 84 HCC patients following liver resection), and found that PANDA was down-regulated in HCC. Thereafter we explored its function in cancer biology by inversing its low expression. Surprisingly, overexpression of PANDA promoted HCC proliferation and carcinogenesis in vitro and in vivo. Mechanistically, PANDA repressed transcriptional activity of senescence associated inflammatory factor IL8, which leaded to inhibition of cellular senescence. Therefore, our research help to better understand the complex role of PANDA in HCC, and suggest more thoughtful strategies should be applied before it can be treated as a potential therapeutic target.
Similar content being viewed by others
Introduction
Hepatocellular carcinoma (HCC) is the second leading cause of cancer death worldwide1. There was about 466 100 newly diagnosed HCC cases and 422 100 patients died from HCC in CHINA in 20152. Although early stage HCC patients benefit from surgical intervention, such as hepatic resection and liver transplantation3, more than half of these patients suffered from recurrence and metastasis after operation. Even worse for advanced HCC patients, so far there is no effective systemic therapy improving overall survival more than 3 months4. Therefore, novel molecular biomarkers for early diagnosis, prognosis evaluation and pharmaceutical options of HCC are extremely important.
Long non-coding RNAs (lncRNAs), as majority of human transcriptome, are a class of RNAs longer than 200 nucleotides that do not encode proteins, and are involved in varieties of biological processes, including cancer biology5,6,7. Long non-coding RNA PANDA (P21 associated ncRNA DNA damage activated, hereafter termed PANDA), is a 5′-capped and polyadenylated non-spliced lncRNA, which located approximately 5 kb upstream of the CDKN1A and transcribed antisense to CDKN1A8. By acting as a molecular decoy9, PANDA interacts with transcription factor NF-YA to suppress its pro-apoptotic function during a DNA damage response8. Meanwhile, PANDA coordinates with scaffold-attachment-factor A (SAFA) to regulate cell senescence and proliferation10.
However, the expression levels and biological functions of PANDA in diverse cancers remain contradictory. It is reported that PANDA was down-regulated in non-small cell lung cancer11 and clear cell renal cell carcinoma12, but up-regulated in gastric cancer13 and breast cancer14. Controversy exits even for the same tissue type of cancer. Puvvula et al.10 reported that PANDA was significantly reduced in HCC, nevertheless Peng et al.15 uncovered that PANDA was overexpressed in HCC. These studies indicate that PANDA plays a complicated role in cancers, and its exact expression level and underlying function in HCC need to be further clearly illuminated.
In the present study, we demonstrated that PANDA was significantly lower expressed in HCC tissue, but conversely had a tumor-promoting feature both in vivo and vitro. Importantly, this aberrant function was attributed to its inhibitory action on cell senescence by suppressing senescence associated inflammatory chemokine IL8.
Results
Low expression of long noncoding RNA PANDA in hepatocellular carcinoma
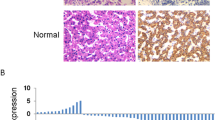
The expression of long noncoding RNA PANDA in hepatocellular carcinoma (HCC) was not clear. Puvvula et al.10 reported that PANDA was low expressed in HCC compared with normal liver tissues and not associated with p53 mutational status in HCC, however, Peng et al.15 uncovered that PANDA was high expressed in HCC tissues and cell lines. Since their data were inconsistent, we employed large collections of clinical samples to further detect PANDA expression by qRT-PCR, cohort one (48 pairs of liver tumor and corresponding peri-tumor samples following liver transplantation) and cohort two (84 pairs of those following hepatic resection). We found that PANDA was significantly low expressed in tumor tissues compared with peri-tumor tissues, p < 0.01 in cohort one (Fig. 1a) and p < 0.001 in cohort two (Fig. 1b). Hung et al.8 reported that PANDA is a p53 effector. To demonstrate whether PANDA was induced by p53-mediated DNA damage in HCC, we treated two liver cancer cell lines HCC-LM3 and Huh7 with doxorubicin (DOX) at 100 nM and 500 nM for 24 hours, and found that PANDA expression was elevated and had a dose-dependent relation(Fig. 1c). Therefore, clinical investigation in our center supported that PANDA was lower expressed in HCC and induced by p53-mediated DNA damage.
PANDA expression level in indicated samples and treatment. Long non-coding RNA PANDA was significantly lower in liver tumor tissue compared with those in peri-tumor tissue in (a) cohort one following liver transplantation and (b) cohort two following liver resection. PANDA was induced in HCC LM3 and Huh7 cell lines when these cells were treated with doxorubicin (DOX) at indicated dose for 24 hours (c).
Overexpression of PANDA promotes hepatocellular carcinoma in vitro
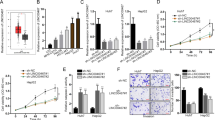
Since the lower expression of PANDA was detected in HCC tissues, we built two highly PANDA-expressing liver cancer cell lines, HCC LM3 and Huh7 by lentivirus (Fig. 2a). PANDA is transcribed antisense to CDKN1A (p21) and located in upstream of CDKN1A8, and CDKN1A functions as a regulator of cell cycle progression, therefore we set CCK-8 assay and EdU assay to evaluate PANDA’s role in cancer cell viability and proliferation. We uncovered that liver cancer cells with higher level of PANDA had an obvious tendency to proliferate. At 24, 48 and 72 hours, HCC LM3 (Fig. 2b) and Huh7 (Fig. 2c) with overexpression of PANDA showed higher OD value than control group. Moreover, HCC LM3 (Fig. 2d) and Huh7 (Fig. 2f) containing higher level of PANDA had more percentage of EdU positive cells, shown in (Fig. 2e,g) respectively. These results indicate that PANDA may have a positive biological function in tumor cell proliferation.
Overexpression of PANDA increases proliferating capacity of liver cancer cells in vitro. (a) Effects of overexpression of PANDA in HCC LM3 and Huh7 cells by lentivirus were verified by qRT-PCR. For CCK-8 assay, HCC LM3 (b) and Huh7 (c) cells with higher expressions of PANDA had significantly elevated OD values (450 nm) at 24, 48, 72 hours. For EdU assay, HCC LM3 (e) and Huh7 (g) cells with higher expressions of PANDA had more percentage of EdU positive cells. Representative pictures are presented in (d,f).
Elevated expression of PANDA augments in vivo carcinogenesis of HCC
To further explore the promoting function of PANDA in tumor proliferation, we injected tumor cells subcutaneously into flanks of immunodeficient mice to build xenograft tumor model. Tumor sizes were measured every four days from the 12th day after implantation. Four weeks later, we sacrificed all the mice and excised tumors out. The representative pictures were shown in Fig. 3a and b. Consistently mice bearing tumor cells with higher level of PANDA had larger tumors (Fig. 3c and d). Then we detected the proliferating activities of tumor masses by staining ki67. The tumor masses generated by highly PANDA-expressing cells had more positive staining areas and stronger intensities of Ki67 (shown in Fig. 3e and analyzed in Fig. 3f). These results supported that long non-coding RNA PANDA not only promoted tumor proliferation in vitro, but also increased tumor growth in vivo.
Overexpression of PANDA promotes liver cancer tumorigenesis in vivo. Mice bearing HCC LM3 (c) cells and Huh7 (d) cells with higher expression of PANDA (OE) had larger xenograft tumor sizes compared with control group (NULL). Representative pictures of 3 specimens from both groups were shown in (a,b) respectively. IHC staining of proliferation marker, Ki67 was consistently stronger in OE group than NULL group (e). IHC staining scores were analyzed in (f).
PANDA reinforces tumor growth by impairing senescence
To illustrate why PANDA could promote liver cancer growth, we used flow cytometry and western blots to assess the impact of PANDA on cell cycle. Unfortunately, no significant transition of G1 phase/S phase in cell cycle has been detected in over-expressing PANDA liver cancer cells compared with control groups, as shown in Fig. 4a. Meanwhile, cell cycle promoting cyclins (Cyclin A, Cyclin D and Cyclin E) and cyclin dependent kinases (CDK1, CDK2, CDK4 and CDK6) were comparative in immunoblotting assay (Fig. 4b). Interestingly, although PANDA was a p53 effector that sensitize tumors to apoptosis by DNA damage8, over-expression of PANDA did not alter p53 expression without external stimulus (Fig. 4b). Moreover, p21, the transcriptional target of p53 which could cause cell cycle arrest, did not change either (Fig. 4b). In proliferating liver cancer cells, HCC LM3 and Huh7, the transcription factor NF-YA remained relatively high, and unchanged by excessive PANDA (Fig. 4b). All above results suggested that PANDA might promote tumor growth through other mechanisms rather than regulation of cell cycle and apoptosis.
PANDA contributes to tumor growth by inhibiting senescence. (a) Representative pictures of cell cycle analysis show that there are no significant difference of G1/S transition between PANDA-overexpressing cells (OE) and control cells (NULL). (b) Cell cycle related proteins in both NULL and OE groups were detected by western blots. The corresponding full-length blots were exhibited in Supplementary Figure 1. (c) In vitro cultured cells stained with senescence associated β-galactosidase in both groups were presented and analyzed statistically. (d) In vivo xenograft tumor samples stained with senescence associated β-galactosidase in both groups were presented and analyzed statistically. (e) Senescence markers p16 and γ-H2AX were detected by western blots in indicated cells. The corresponding full-length blots were exhibited in Supplementary Figure 2. (f) Senescence markers p16 and γ-H2AX were detected by IHC in indicated xenograft tumor samples. (g) IHC staining scores were analyzed.
It has been reported that PANDA recruited complexes to repress senescence-promoting genes10. Positive blue signal in β-galactosidase staining was a biomarker that well identified in senescent cells both in culture and in vivo 16. We employed β-galactosidase staining to identify senescent cells in PANDA-overexpressing groups and control groups. Even though the number of senescent cells among liver cancer cells was relatively low, there were significantly fewer blue stained cells in OE groups than NULL groups (Fig. 4c). And for xenograft tumor nodules, in vivo β-galactosidase staining showed similar results (Fig. 4d). Moreover, other markers of celluar senescence, such as cell cycle inhibitor p1617 and senescence-associated DNA damage foci characterized by phosphorylated histone H2AX (γ-H2AX)18, were examined in cultured cell lines and xenograft tumor samples. We also found that p16 and γ-H2AX were decreased while PANDA was overexpressed (Fig. 4e–g). These results indicated that overexpression of PANDA impaired senescence in tumor cells, and these tumor cells who bypassed senescence barrier contributed to tumor growth.
Over expression of PANDA inhibits senescence associated factor IL8
The molecular mechanism that lncRNA PANDA regulates cellular processes was to bind to transcription factors or RNA binding proteins, such as NF-YA or SAFA, and then to form polycomb repressive complexes (PRCs) to regulate downstream genes8, 10. DUSP4, JUNB, IL8, MCM3, PCNA, LB1, TK1 had been reported as PRCs’ target genes and involved in cellular proliferation and senescence10. In order to determine whether these target genes were modulated by PANDA in our research, we used qRT-PCR to examine their expression. Remarkably, only chemokine IL8 was restrained to half level in PANDA-overexpressing group compared with control group both in HCC LM3 and Huh7(Fig. 5a). IL8 was a typical chemokine secreted by senescent cells and induced neighboring cells to occur senescence by paracrine fashion19. We hypothesized that redundant PANDA inactivated the IL8 transcription, caused less IL8 secretion, thus inhibited cellular senescence. To further verify this hypothesis, we investigated IL8 secretion from cell culture supernatant using ELISA, and IL8 staining from xenograft samples using immunohistochemistry. Consistently, cells expressing higher PANDA released less IL8 into cell culture (Fig. 5b), and tumor nodules generated from highly-expressing-PANDA cells expressed lower IL8 (Fig. 5c and d). In addition, clinically we examined these PRC target genes as well as PANDA in a small number of HCC tumor samples from cohort two, and analyzed their correlation. Among all these target genes, IL8 had a significant negative relevance with PANDA (Fig. 5e). Therefore, we discovered that IL8 was downregulated by lncRNA PANDA during cellular proliferation and senescence.
Overexpression of PANDA inhibits senescence associated factor IL8. (a) The transcriptional target genes of PANDA-PRC complexes were examined by qRT-PCR in both PANDA-overexpressing groups (OE) and control groups (NULL). (b) Cells in OE group secreted less IL8 into culture supernatant comparing with NULL group. (c) IL8 staining in xenograft model samples generated by OE cells were stronger than those of NULL cells. (d) IHC staining scores were analyzed. (e) The clinical relevance of PRC target genes and lncRNA PANDA were analyzed in a small number of patients in cohort two.
Ectopic expression of IL8 rescues PANDA-mediated phenotype
In order to demonstrate the enhanced proliferation and bypassed senescence in PANDA-overexpressed cells is participated by IL8, we induced IL8 expression by plasmids in Huh7-PANDA-OE and HCC LM3-PANDA-OE cells (Fig. 6a). We found that overexpression of IL8 in these cells could reverse the phenotype caused by introduction of PANDA, detected by CCK-8 assay (Fig. 6b), EdU assay (Fig. 6c) and in vitro β-galactosidase staining (Fig. 6d). Oncogene-induced senescence was a classical model to dissect senescence19. We transfected BJ fibroblast cells with NRasG12V plasmids, which had been used for senescence induction20, to further confirm that lncRNA PANDA and IL8 had a negative relevance during senescence. From the fifth day to the tenth day after transfection, IL8 increased while PANDA decreased (Fig. 6e). These results indicated that IL8 mediated the biological function of PANDA in HCC.
Overexpression of IL8 rescues PANDA-mediated phenotype. (a) Effects of overexpression of IL8 in HCC LM3-PANDA-OE and Huh7-PANDA-OE cells by plasmid were confirmed by western blots. The corresponding full-length blots were exhibited in Supplementary Figure 3. (b) PANDA-overexpressed cells transfected with IL8 plasmid had reduced OD values (450 nm) at 24, 48, 72 hours compared with those with empty vector. (c) For EdU assay, PANDA-overexpressed cells transfected with IL8 plasmid had less percentage of EdU positive cells. (d) For in vitro senescence associated β-galactosidase staining, PANDA-overexpressed cells transfected with IL8 plasmid had more blue stained cells. (e) RNA expression levels of PANDA and IL8 were analyzed in BJ fibroblasts at indicated time after transfected with NRasG12V plasmid. (f) The reporter activity of IL8 promoter in 293 T with or without overexpression of PANDA was determined by dual luciferase reporter assay system. (g) Huh7 cell lysates were incubated with in vitro-transcribed biotin-labeled sense or antisense PANDA for RNA pull-down assay, followed by western blot analysis with indicated antibodies. The corresponding full-length blots were exhibited in Supplementary Figure 4.
However, the underlying mechanism how PANDA regulated IL8 still remained unknown. We performed a dual luciferase reporter assay in 293 T cells to evaluate the reporter activity of IL8 promoter with or without overexpression of PANDA. As a result, the reporter activity of IL8 promoter decreased significantly when transfected with PANDA plasmid (Fig. 6f). To demonstrate whether lncRNA PANDA directly bound to IL8 protein, we conducted RNA pull down assay. Unfortunately, no evidence of direct binding of lncRNA PANDA and protein IL8 could be observed (Fig. 6g). Therefore, lncRNA PANDA might inhibit IL8 expression by forming a polycomb repressive complex targeting IL8 promoter, rather than directly binding to IL8 protein.
Discussion
Recently, long non-coding RNA PANDA has been examined in more and more types of cancers, but its expression level and biological function has tissue specific property and remains controversial10,11,12,13,14,15. The inconsistent results of PANDA expression in HCC between Puvvula et al.’s research10 and Peng et al.’s research15 attracted our considerable interests. In our study, PANDA was significantly reduced in two independent cohorts of HCC patient samples. And PANDA was first reported in the literature as a effector of tumor suppressor p53, and loss of PANDA made cells more sensitive to apoptosis8. Therefore, in combination with our results and literature reviews, we strongly supported that PANDA was down-regulated in HCC. However, PANDA expression pattern in HCC still needs to be more clearly defined by multicentre evaluation and validation, especially according to the p53 status.
Unexpectedly, we observed a tumor promoting function of PANDA in HCC through the methods of overexpressing PANDA by lentivirus. PANDA not only increased tumor cell proliferation in cell culture but also accelerated carcinogenesis in xenograft tumor model. Oncogenes or tumor suppressors played unexpected tumor-suppressing or tumor-promoting roles in hepatocarcinogenesis had driven increasing attention recently21. He et al.22, Bard-Chapeau et al.23. and Wang et al.24. had explained that the conflicting phenotypes may be caused by liver compensatory proliferation due to alteration of inflammatory microenvironment. These inspired us to search underlying mechanism, not limited to cell cycle transition and apoptosis. Interestingly, we uncovered that the cellular senescence of liver cancer cells was inhibited by elevated level of PANDA.
Cellular senescence is a biological process that cell responses to diverse stress, including dysfunctional telomeres, DNA damage and strong mitogenic signals, thus acquires a long term state of proliferate arrest25. Senescent cells can drive degenerative and hyperplastic pathology, the most deadly of the latter is cancer26. Senescence-to-immortality transition has been revealed in hepatocytes, which is mediated by genome-wide transcriptional modifications involved with long non-coding RNAs27. One hallmark feature of senescence is that senescent cells express and secrete cytokines and growth factors to increase the complexity of tumor microenvironment, which is termed senescence-associated secretory phenotype (SASP), including IL-6, IL-828. These senescence associated factors have considerable influence on neighbouring cells through paracrine fashion to apply beneficial or detrimental effects19.
PANDA inhibits senescence-promoting genes in proliferating cancerous cells by transcriptional interference10. We detected PANDA target genes and found that IL8 was significantly reduced when PANDA is overexpressed. The senescence associated inflammatory mediator, IL8 was produced when transcription factor NF-κB was activated, and its expression can be promoted by protein kinase D1 (PKD1)29 and suppressed by microRNAs miR-146a/b30. Induction of IL8 occurs in cells undergoing senescence, while repression of IL8 delays replicative senescence in outgrowth endothelial cells31. Here, we demonstrated that IL8 was suppressed by PANDA in HCC, thereafter inhibited cellular senescence, which allowed pre-senescent cells to escape senescence, and continue to proliferate, thus contribute to tumor promotion.
The molecular mechanism that employed by lncRNAs to regulate cellular processes has been well stated32. Based on the fact that the cellular location of lncRNA PANDA was nuclear, PANDA functionally participated in chromatin interactions and transcriptional regulation7. It is in agreement with our findings. We revealed that overexpression of PANDA directly decrease the transcription activity of IL8 promoter, but not directly binding to IL8 protein. However, how the transcription activity of IL8 promoter were suppressed by PANDA, through facilitating histone modification or serving as a assembling scaffold, remains undiscovered.
In summary, we expanded our knowledge about long non-coding RNA PANDA in HCC that PANDA was downregulated in HCC but promoted liver cancer tumorigenesis by inhibiting cellular senescence via senescence associated inflammatory mediator IL8. Our findings provide a profound understanding about the complicated role of long non-coding RNA PANDA in liver cancer, and emphasize that precise molecular mechanisms should be elucidated before targeting it as diagnostic and therapeutic agents. Moreover, we strongly suggest that genetically engineered animal model or different p53 status model should be required to investigate the biological function of lncRNA PANDA in cancer.
Methods
Patients and samples
48 frozen HCC tissues and the corresponding peri-tumor tissues were obtained from patients undergoing liver transplantation at the First Affiliated Hospital, School of Medicine, Zhejiang University (Hangzhou, China) between 2011 and 2012 (cohort one). Additional 84 HCC tissues were collected from patients following liver resection at the First Affiliated Hospital, School of Medicine, Zhejiang University between 2010 and 2013 (cohort two). Informed consents have been obtained from all patients. And the research was conducted with the approval of Institutional Review Board and Ethics Committee of First Affiliated Hospital, School of Medicine, Zhejiang University, and in accordance with Declaration of Helsinki.
RNA extraction and qRT-PCR analysis
Total RNA from the specimens and cells was isolated using Trizol reagent (Invitrogen, Carlsbad, California, USA). Reverse Transcription was performed using High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Waltham, Massachusetts, USA) and qRT-PCR was performed with SYBR® Green dye (Takara, Shiga, Japan). All reactions were performed in triplicate and GAPDH RNA levels were used for normalization. The primer sequences were provided in Supplementary Table S1.
Cell culture
Hepatocellular carcinoma cell lines, HCC LM3 and Huh7, were purchased from China Center for Type Culture Collection (CCTCC), and grown in MEM supplemented with 10% fetal bovine serum (FBS) and penicillin/streptomycin, at 37°C under an atmosphere containing 5% CO2. For doxorubicin (DOX, Sigma D1515, St. Louis, Missouri, USA) treatment, DOX was added to culture medium at 0 nM, 100 nM, 500 nM and maintained for 24 hours.
Overexpression of PANDA and IL8
The lentiviral PANDA construct (OE) and empty construct (NULL) were completed by Genechem Company (Shanghai, China). Full-length PANDA (NR_109836) was gene synthesized, sequence-verified and cloned into GV341 (Ubi-MCS-3FLAG-SV40-puromycin) vector (Genechem, Shanghai, China). Lentiviral infection was performed by standard procedures. The IL8 plasmid and empty vector were purchased from Genechem Company (Shanghai, China). Transfection was performed with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instructions.
Cell Proliferation Assay
5 × 103 cells were seeded into each well of 96-well plate to perform Cell Counting Kit-8 (CCK-8) proliferation assay (Dojindo, Kumamoto, Japan) at indicated time according to the manufacturer’s instructions as previously described33. 2 × 104 cells were seeded into Nunc™ Glass Bottom Dishes (Thermo Scientific, 150682, Waltham, Massachusetts, USA) to complete EdU imaging assay (RiboBio, C10310, Guangzhou, China) after 24 hours according to the manufacturer’s instructions.
Immunoblotting analysis and antibodies
Protein extraction and immunoblotting analysis were performed according to the regular procedure. The following antibodies were used: NF-YA (Santa Cruz, sc-10779, Dallas, Texas, USA), CDK1 (CST, #9116, Danvers, Massachusetts, USA), CDK2 (CST, #2546), CDK4 (CST, #12790), CDK6 (CST, #3136), Cyclin A (CST, #4656), Cyclin D (CST, #2978), Cyclin E (CST, #4129), p53 (Santa Cruz, sc-126), p21 (CST, #2947), b-actin (CST, #8457), β-tubulin (CST, #2128), p16 (proteintech, 10883-1-AP), γ-H2AX (abcam, ab81299) and IL8 (ABGENT, AP8612B, San Diego, California, USA). The original full-length blots were exhibited in the Supplementary Information.
Xenograft model and immunohistochemistry
5 × 106 HCC LM3 and Huh7 cells expressing control (NULL) or PANDA (OE) were suspended in 0.2 ml PBS and injected subcutaneously into flanks of 8 weeks old immunodeficient mice. Tumor volumes were measured every four days from the 12th day after injection, and calculated by the formula V = ½ (length × width2). All the mice were sacrificed 4 weeks later after injection and tumors were harvested for immunostaining. Animal study protocols were performed in accordance with the National Institute Guide for the care and use of laboratory animals.
Immunohistochemistry and semiquantitative evaluation were performed as described previously24. Antibodies recognizing Ki67 (abcam, ab15580, Cambridge, UK), IL8 (ABGENT, AP8612B, San Diego, California, USA), p16 (proteintech, 10883-1-AP, Wuhan, P.R.C.) and γ-H2AX (abcam, ab81299, Cambridge, UK) were used in immunostaining.
Senescence analysis
Senescence was assessed by β-galactosidase cell staining according to manufacturer’s instructions (CST, #9860). For in vitro assay, 1 × 104 cells were seeded into Nunc™ Glass Bottom Dishes (Thermo Scientific, 150682, Waltham, Massachusetts, USA) and then followed by standard procedure. For in vivo assay, xenograft tumors were fixed with 0.2% PFA/0.1 M PIPES/2 mM MgCl2/5 mM EGTA (pH = 6.0) before freezing and then followed by standard procedure.
Cell cycle analysis
Cells were stained with propidium iodide (BD Biosciences, San Jose, California, USA), after 75% ethanol fixation overnight, and analyzed for DNA content by flow cytometry (BD Biosciences) according to manufacturer’s instructions. The data were analyzed using ModFit LTTM (Verity Software, Topsham, Maine, USA).
ELISA
3 × 105 cells were seeded into each well of 6-well plate. After serum starvation for 24 hours, cell culture supertatant was collected for detection of IL8 by Human IL-8 Platinum ELISA (BMS204/3, eBioscience, San Diego, California, USA) according to manufacturer’s instructions.
Luciferase report assay
The 2 kb IL8 promoter upstream of the starting codon ATG was synthesized and cloned into pGL3-basic plasmid by Genechem Company (Shanghai, China). The reporter activities were measured 48 hours after co-transfection using the Dual-Luciferase Reporter Assay System (Promega, E1910). After normalization, the ratios of Firefly/Renilla luminescence were presented.
RNA pull down assay
In vitro transcription was performed by using T7 RNA polymerase (Roche, 10881767001), SP6 RNA polymerase (Roche, 10810274001) and Biotin RNA Labeling Mix (Roche, 11685597910) according to the instructions provided by the manufacturer. 3 ug in vitro-transcribed biotin-labeled PANDA was incubated with 1 mg Huh7 lysates for 1 hour at 4 °C. Streptavidin-coupled Dynabeads M-280 (Invitrogen, 11205D) were then added to the reaction mix to isolate the RNA-protein complex.
Statistical analysis
Statistical analyses were performed using Microsoft Office Excel and GraphPad Prism 6. Data were presented as the mean ± SD, and differences between groups were evaluated by two-tailed independent Student’s t-test. Relevance analysis was performed with Pearson’s correlation. P value less than 0.5 was recognized as statistically significant. All experiments were performed at least three replicates or three independent assays. *p < 0.05. **p < 0.01. ***p < 0.001.
References
Torre, L. A. et al. Global cancer statistics, 2012. CA Cancer J Clin. 65, 87–108 (2015).
Chen, W. et al. Cancer statistics in China, 2015. CA Cancer J Clin. 66, 115–32 (2016).
Llovet, J. M., Burroughs, A. & Bruix, J. Hepatocellular carcinoma. Lancet. 362, 1907–17 (2003).
Llovet, J. M. et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 359, 378–90 (2008).
Djebali, S. et al. Landscape of transcription in human cells. Nature. 489, 101–8 (2012).
Quinn, J. J. & Chang, H. Y. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 17, 47–62 (2016).
Schmitt, A. M. & Chang, H. Y. Long Noncoding RNAs in Cancer Pathways. Cancer Cell. 29, 452–63 (2016).
Hung, T. et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet. 43, 621–9 (2011).
He, Y. et al. Long noncoding RNAs: Novel insights into hepatocelluar carcinoma. Cancer Lett. 344, 20–7 (2014).
Puvvula, P. K. et al. Long noncoding RNA PANDA and scaffold-attachment-factor SAFA control senescence entry and exit. Nat Commun. 5, 5323 (2014).
Han, L. et al. Low expression of long noncoding RNA PANDAR predicts a poor prognosis of non-small cell lung cancer and affects cell apoptosis by regulating Bcl-2. Cell Death Dis. 6, e1665 (2015).
Wu, Y. et al. A serum-circulating long noncoding RNA signature can discriminate between patients with clear cell renal cell carcinoma and healthy controls. Oncogenesis. 5, e192 (2016).
Ma, P., Xu, T., Huang, M. & Shu, Y. Increased expression of LncRNA PANDAR predicts a poor prognosis in gastric cancer. Biomed Pharmacother. 78, 172–6 (2016).
Sang, Y. et al. LncRNA PANDAR regulates the G1/S transition of breast cancer cells by suppressing p16(INK4A) expression. Sci Rep. 6, 22366 (2016).
Peng, W. & Fan, H. Long non-coding RNA PANDAR correlates with poor prognosis and promotes tumorigenesis in hepatocellular carcinoma. Biomed Pharmacother. 72, 113–8 (2015).
Dimri, G. P. et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci USA 92, 9363–7 (1995).
Campisi, J. Replicative senescence: an old lives’ tale? Cell. 84, 497–500 (1996).
d’Adda di Fagagna, F. et al. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 426, 194–8 (2003).
Aravinthan, A. D. & Alexander, G. J. Senescence in chronic liver disease: Is the future in aging? J Hepatol. 65, 825–34 (2016).
Eggert, T. et al. Distinct Functions of Senescence-Associated Immune Responses in Liver Tumor Surveillance and Tumor Progression. Cancer Cell. 30, 533–547 (2016).
Feng, G. S. Conflicting roles of molecules in hepatocarcinogenesis: paradigm or paradox. Cancer Cell. 21, 150–4 (2012).
He, G. et al. Hepatocyte IKKbeta/NF-kappaB inhibits tumor promotion and progression by preventing oxidative stress-driven STAT3 activation. Cancer Cell. 17, 286–97 (2010).
Bard-Chapeau, E. A. et al. Ptpn11/Shp2 acts as a tumor suppressor in hepatocellular carcinogenesis. Cancer Cell. 19, 629–39 (2011).
Wang, Q. et al. Spontaneous Hepatocellular Carcinoma after the Combined Deletion of Akt Isoforms. Cancer Cell. 29, 523–35 (2016).
Campisi, J. Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell. 120, 513–22 (2005).
Judith, C. A. Cellular Senescence, and Cancer. Annu Rev Physiol. 75, 685–705 (2013).
Yildiz, G. et al. Genome-wide transcriptional reorganization associated with senescence-to-immortality switch during human hepatocellular carcinogenesis. PLoS One. 8, e64016, doi:10.1371/journal.pone.0064016 (2013).
Abdelmohsen, K. & Gorospe, M. Noncoding RNA control of cellular senescence. Wiley Interdiscip Rev RNA. 6, 615–29 (2015).
Wang, P. et al. Protein kinase D1 is essential for Ras-induced senescence and tumor suppression by regulating senescence-associated inflammation. Proc Natl Acad Sci USA 111, 7683–8 (2014).
Bhaumik, D. et al. MicroRNAs miR-146a/b negatively modulate the senescence-associated inflammatory mediators IL-6 and IL-8. Aging (Albany NY). 1, 402–11 (2009).
Medina, R. J. et al. Ex vivo expansion of human outgrowth endothelial cells leads to IL-8-mediated replicative senescence and impaired vasoreparative function. Stem Cells. 31, 1657–68 (2013).
Guttman, M. & Rinn, J. L. Modular regulatory principles of large non-coding RNAs. Nature. 482, 339–46 (2012).
Peng, C. et al. A critical role for ZDHHC2 in metastasis and recurrence in human hepatocellular carcinoma. Biomed Res Int. 2014, 832712 (2014).
Acknowledgements
This study was supported by the Major Research plan of the National Natural Science Foundation of China (No.91542205), the National Health and Family Planning Commission of China (No.2016138643), the Science Technology Department of Zhejiang Province (No.2015C03034), the Zhejiang Provincial Natural Science Foundation (No.LY15H160016 and No.LQ15H160002) and the General Project Plan of Zhejiang Medical Technology (No.2016148875).
Author information
Authors and Affiliations
Contributions
C.P., J.W. and S.Z. conceived the experiments. C.P., W.H., X.W. and R.T. conducted the experiments. S.C. and C.D. evaluated the histological score. H.X., Z.L., H.X. and L.Z. analyzed the results. C.P., J.W. and S.Z. drafted the manuscript. C.P. and Z.X. prepared the revised article. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Peng, C., Hu, W., Weng, X. et al. Over Expression of Long Non-Coding RNA PANDA Promotes Hepatocellular Carcinoma by Inhibiting Senescence Associated Inflammatory Factor IL8. Sci Rep 7, 4186 (2017). https://doi.org/10.1038/s41598-017-04045-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-04045-5
This article is cited by
-
A novel aging-associated lncRNA signature for predicting prognosis in osteosarcoma
Scientific Reports (2024)
-
LINC00355 regulates p27KIP expression by binding to MENIN to induce proliferation in late-stage relapse breast cancer
npj Breast Cancer (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.