Abstract
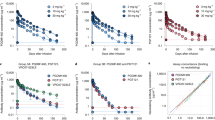
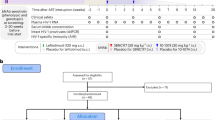
Monotherapy of HIV-1 infection with single antiretroviral agents is ineffective because error-prone HIV-1 replication leads to the production of drug-resistant viral variants1,2. Combinations of drugs can establish long-term control, however, antiretroviral therapy (ART) requires daily dosing, can cause side effects and does not eradicate the infection3,4. Although anti-HIV-1 antibodies constitute a potential alternative to ART5,6, treatment of viremic individuals with a single antibody also results in emergence of resistant viral variants7,8,9. Moreover, combinations of first-generation anti-HIV-1 broadly neutralizing antibodies (bNAbs) had little measurable effect on the infection10,11,12. Here we report on a phase 1b clinical trial (NCT02825797) in which two potent bNAbs, 3BNC11713 and 10-107414, were administered in combination to seven HIV-1 viremic individuals. Infusions of 30 mg kg−1 of each of the antibodies were well-tolerated. In the four individuals with dual antibody-sensitive viruses, immunotherapy resulted in an average reduction in HIV-1 viral load of 2.05 log10 copies per ml that remained significantly reduced for three months following the first of up to three infusions. In addition, none of these individuals developed resistance to both antibodies. Larger studies will be necessary to confirm the efficacy of antibody combinations in reducing HIV-1 viremia and limiting the emergence of resistant viral variants.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All requests for raw and analyzed data and materials are promptly reviewed by the Rockefeller University to verify whether the request is subject to any intellectual property or confidentiality obligations. Patient-related data not included in the paper were generated as part of clinical trials and may be subject to patient confidentiality. Any data and materials that can be shared will be released via a Material Transfer Agreement. HIV-1 envelope SGA data are available in GenBank, accession numbers MH632763–MH633255.
References
Ndung’u, T. & Weiss, R. A. On HIV diversity. AIDS 26, 1255–1260 (2012).
Bailey, J., Blankson, J. N., Wind-Rotolo, M. & Siliciano, R. F. Mechanisms of HIV-1 escape from immune responses and antiretroviral drugs. Curr. Opin. Immunol. 16, 470–476 (2004).
Siliciano, J. D. et al. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat. Med. 9, 727–728 (2003).
Finzi, D. et al. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat. Med. 5, 512–517 (1999).
Walker, L. M. & Burton, D. R. Passive immunotherapy of viral infections: ‘super-antibodies’ enter the fray. Nat. Rev. Immunol. 18, 297–308 (2018).
Klein, F. et al. Antibodies in HIV-1 vaccine development and therapy. Science 341, 1199–1204 (2013).
Caskey, M. et al. Viraemia suppressed in HIV-1-infected humans by broadly neutralizing antibody 3BNC117. Nature 522, 487–491 (2015).
Caskey, M. et al. Antibody 10-1074 suppresses viremia in HIV-1-infected individuals. Nat. Med. 23, 185–191 (2017).
Lynch, R. M. et al. Virologic effects of broadly neutralizing antibody VRC01 administration during chronic HIV-1 infection. Sci. Transl. Med. 7, 319ra206 (2015).
Armbruster, C. et al. Passive immunization with the anti-HIV-1 human monoclonal antibody (hMAb) 4E10 and the hMAb combination 4E10/2F5/2G12. J. Antimicrob. Chemother. 54, 915–920 (2004).
Mehandru, S. et al. Adjunctive passive immunotherapy in human immunodeficiency virus type 1-infected individuals treated with antiviral therapy during acute and early infection. J. Virol. 81, 11016–11031 (2007).
Trkola, A. et al. Delay of HIV-1 rebound after cessation of antiretroviral therapy through passive transfer of human neutralizing antibodies. Nat. Med. 11, 615–622 (2005).
Scheid, J. F. et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science 333, 1633–1637 (2011).
Mouquet, H. et al. Complex-type N-glycan recognition by potent broadly neutralizing HIV antibodies. Proc. Natl Acad. Sci. USA 109, E3268–E3277 (2012).
Mendoza, P. et al. Combination therapy with anti-HIV-1 antibodies maintains viral suppression. Nature https://doi.org/10.1038/s41586-018-0531-2 (2018).
Sarzotti-Kelsoe, M. et al. Optimization and validation of the TZM-bl assay for standardized assessments of neutralizing antibodies against HIV-1. J. Immunol. Methods 409, 131–146 (2014).
Scheid, J. F. et al. HIV-1 antibody 3BNC117 suppresses viral rebound in humans during treatment interruption. Nature 535, 556–560 (2016).
Cohen, Y. Z. et al. Neutralizing activity of broadly neutralizing anti-HIV-1 antibodies against clade B clinical isolates produced in peripheral blood mononuclear cells. J. Virol. 92, e01883-17 (2018).
Keizer, R. J., Huitema, A. D., Schellens, J. H. & Beijnen, J. H. Clinical pharmacokinetics of therapeutic monoclonal antibodies. Clin. Pharmacokinet. 49, 493–507 (2010).
Horwitz, J. A. et al. HIV-1 suppression and durable control by combining single broadly neutralizing antibodies and antiretroviral drugs in humanized mice. Proc. Natl Acad. Sci. USA 110, 16538–16543 (2013).
Klein, F. et al. HIV therapy by a combination of broadly neutralizing antibodies in humanized mice. Nature 492, 118–122 (2012).
Klein, F. et al. Enhanced HIV-1 immunotherapy by commonly arising antibodies that target virus escape variants. J. Exp. Med. 211, 2361–2372 (2014).
Shingai, M. et al. Antibody-mediated immunotherapy of macaques chronically infected with SHIV suppresses viraemia. Nature 503, 277–280 (2013).
Nishimura, Y. et al. Early antibody therapy can induce long-lasting immunity to SHIV. Nature 543, 559–563 (2017).
Walker, B. D. & Yu, X. G. Unravelling the mechanisms of durable control of HIV-1. Nat. Rev. Immunol. 13, 487–498 (2013).
Lynch, R. M. et al. HIV-1 fitness cost associated with escape from the VRC01 class of CD4 binding site neutralizing antibodies. J. Virol. 89, 4201–4213 (2015).
Diskin, R. et al. Restricting HIV-1 pathways for escape using rationally designed anti-HIV-1 antibodies. J. Exp. Med. 210, 1235–1249 (2013).
Gaudinski, M. R. et al. Safety and pharmacokinetics of the Fc-modified HIV-1 human monoclonal antibody VRC01LS: a phase 1 open-label clinical trial in healthy adults. PLoS Med. 15, e1002493 (2018).
Ko, S. Y. et al. Enhanced neonatal Fc receptor function improves protection against primate SHIV infection. Nature 514, 642–645 (2014).
Robbie, G. J. et al. A novel investigational Fc-modified humanized monoclonal antibody, motavizumab-YTE, has an extended half-life in healthy adults. Antimicrob. Agents Chemother. 57, 6147–6153 (2013).
Gautam, R. et al. A single injection of crystallizable fragment domain-modified antibodies elicits durable protection from SHIV infection. Nat. Med. 24, 610–616 (2018).
Zhou, T. et al. Multidonor analysis reveals structural elements, genetic determinants, and maturation pathway for HIV-1 neutralization by VRC01-class antibodies. Immunity 39, 245–258 (2013).
Schoofs, T. et al. HIV-1 therapy with monoclonal antibody 3BNC117 elicits host immune responses against HIV-1. Science 352, 997–1001 (2016).
Larkin, M. A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007).
Hraber, P. et al. Longitudinal antigenic sequences and sites from intra-host evolution (LASSIE) identifies immune-selected HIV variants. Viruses 7, 5443–5475 (2015).
Guindon, S. et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321 (2010).
Stamatakis, A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Kirchherr, J. L. et al. High throughput functional analysis of HIV-1 env genes without cloning. J. Virol. Methods 143, 104–111 (2007).
Acknowledgements
We thank all study participants who devoted time to our research; members of the Klein and Nussenzweig laboratories for helpful discussions, especially P. Mendoza, C.-L. Lu, J. C. C. Lorenzi, L. Cohn and M. Jankovic; R. Levin, G. Kremer and D. Weiland for study coordination; the Rockefeller University Hospital Clinical Research Support Office and nursing staff as well as C. Golder, E. Thomas, M. Platten, S. Margane and T. Kümmerle for help with recruitment and study implementation; C. Ruping and M. Schlotz for help with sample processing; S. Kiss for ophthalmologic assessments; T. Keler and the Celldex Therapeutics team for 3BNC117 and 10-1074 manufacturing and regulatory support; C. Conrad for regulatory support; U. Kerkweg for pharmaceutical services; H. Janicki, M. Ercanoglu, P. Schommers and R. Kaiser for help with virus cultures; P. Fast and H. Park for clinical monitoring; S. McMillan, S. Mosher, S. Sawant, D. Beaumont, M. Sarzotti-Kelsoe, K. Greene, H. Gao and D. Montefiori for help with PK assay development, validation, reporting and/or project management; and S. Schlesinger for input on study design. This work was supported by The Bill and Melinda Gates Foundation Collaboration for AIDS Vaccine Discovery (CAVD) grants OPP1092074, OPP1124068 (M.C.N.), CAVIMC OPP1146996 (G.D.T., M.S.S.); the NIH grants 1UM1 AI100663 and R01AI-129795 (M.C.N.); the Heisenberg-Program of the DFG (KL 2389/2-1), the European Research Council (ERC-StG639961) and the German Center for Infection Research (DZIF) (F.K.); the Einstein-Rockefeller-CUNY Center for AIDS Research (1P30AI124414-01A1); BEAT-HIV Delaney grant UM1 AI126620 (M.C.); and the Robertson fund. M.C.N. is a Howard Hughes Medical Institute Investigator.
Author information
Authors and Affiliations
Contributions
M.C. (principal investigator in the United States), F.K. (principal investigator in Germany) and M.C.N. designed the trial; Y.B.-O., H.G., M.C., F.K. and M.C.N. analyzed the data and wrote the manuscript; Y.B.-O., T.S. and T.K. performed single-genome sequencing; H.G., A.L.B., K.M., M.W.-P., K.F., J.H., M.C. and F.K. implemented the study; Y.Z.C., R.M.G. and G.F. contributed to study design and implementation; C.L., I.Su., C.W. and S.S. contributed to participant recruitment and clinical assessments; J.A.P. and T.Y.O. performed bioinformatics processing; H.G., L.N. and T.K. performed viral cultures; L.H. and N.P. contributed to statistical analyses; S.B., J.P.D., J.J.V., I.Sh. and K.J. performed, coordinated or contributed to sample processing; K.E.S., N.L.Y. and G.D.T. performed anti-idiotypic ELISAs; and M.S.S. performed neutralization assays.
Corresponding authors
Ethics declarations
Competing interests
There are patents on 3BNC117 and 10-1074 on which M.C.N. is an inventor.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–9 and Supplementary Tables 1–6
Rights and permissions
About this article
Cite this article
Bar-On, Y., Gruell, H., Schoofs, T. et al. Safety and antiviral activity of combination HIV-1 broadly neutralizing antibodies in viremic individuals. Nat Med 24, 1701–1707 (2018). https://doi.org/10.1038/s41591-018-0186-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41591-018-0186-4
This article is cited by
-
Mucosal application of the broadly neutralizing antibody 10-1074 protects macaques from cell-associated SHIV vaginal exposure
Nature Communications (2023)
-
Probabilities of developing HIV-1 bNAb sequence features in uninfected and chronically infected individuals
Nature Communications (2023)
-
Enhancing anti-viral neutralization response to immunization with HIV-1 envelope glycoprotein immunogens
npj Vaccines (2023)
-
Impact of a TLR9 agonist and broadly neutralizing antibodies on HIV-1 persistence: the randomized phase 2a TITAN trial
Nature Medicine (2023)
-
HIV-1 bispecific antibody iMab-N6 exhibits enhanced breadth but not potency over its parental antibodies iMab and N6
Virology Journal (2022)