Abstract
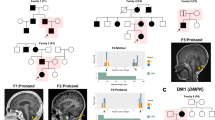
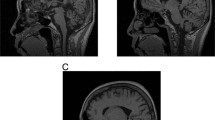
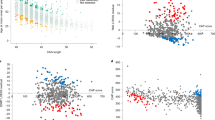
Despite linkage to chromosome 16q in 1996, the mutation causing spinocerebellar ataxia type 4 (SCA4), a late-onset sensory and cerebellar ataxia, remained unknown. Here, using long-read single-strand whole-genome sequencing (LR-GS), we identified a heterozygous GGC-repeat expansion in a large Utah pedigree encoding polyglycine (polyG) in zinc finger homeobox protein 3 (ZFHX3), also known as AT-binding transcription factor 1 (ATBF1). We queried 6,495 genome sequencing datasets and identified the repeat expansion in seven additional pedigrees. Ultrarare DNA variants near the repeat expansion indicate a common distant founder event in Sweden. Intranuclear ZFHX3–p62–ubiquitin aggregates were abundant in SCA4 basis pontis neurons. In fibroblasts and induced pluripotent stem cells, the GGC expansion led to increased ZFHX3 protein levels and abnormal autophagy, which were normalized with small interfering RNA-mediated ZFHX3 knockdown in both cell types. Improving autophagy points to a therapeutic avenue for this novel polyG disease. The coding GGC-repeat expansion in an extremely G+C-rich region was not detectable by short-read whole-exome sequencing, which demonstrates the power of LR-GS for variant discovery.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The data that support the findings of this study are available on request from the corresponding authors (S.M.P. and O.R.). The data are not publicly available as they contain information that could compromise the privacy of research participants. Source data are provided with this paper.
Code availability
(1) Open-source tools and pipelines: (1.1) in-house developed tools and pipelines: megSAP version 2022_08 (short-read WGS data): https://github.com/imgag/megSAP, https://doi.org/10.5281/zenodo.10817663 (ref. 57); megLR v1.0.0 (long-read WGS data): https://github.com/imgag/megLR, https://doi.org/10.5281/zenodo.10820153 (ref. 58); Expander (repeat expansion detection with Nanopore data): https://github.com/caspargross/expander, https://doi.org/10.5281/zenodo.10820172 (ref. 59); SCA4 project-specific scripts (scripts for data formatting, plotting, haplotype analysis and linkage analysis): https://github.com/imgag/sca4_project; (1.2) other open-source tools for long-read WGS analysis (used by in-house pipeline): trgt v0.4.0 (repeat expansion detection with long-read WGS HiFi): https://github.com/PacificBiosciences/trgt; Straglr v1.4.0 (repeat expansion detection with long-read WGS Nanopore): https://github.com/bcgsc/straglr; pbsv v2.9.0 (structural variant detection with long-read WGS HiFi): https://github.com/PacificBiosciences/pbsv; Sniffles2 v2.0.7 (structural variant detection with long-read WGS Nanopore): https://github.com/fritzsedlazeck/Sniffles; minimap2 v2.25 (long-read WGS alignment): https://github.com/lh3/minimap2; Flye v2.9.2 (genome assembly with long-read WGS): https://github.com/fenderglass/Flye; pb-assembly v1 (genome assembly with long-read WGS HiFi): https://github.com/PacificBiosciences/pb-assembly; hapdup v0.12 (haplotype phasing): https://github.com/KolmogorovLab/hapdup; hapdiff v0.8 (phased structural variant calling): https://github.com/KolmogorovLab/hapdiff; PEPPER-Margin-DeepVariant r0.8 (variant calling with long-read WGS Nanopore): https://github.com/kishwarshafin/pepper/releases/tag/r0.8. (2) Vendor software for sequencing platforms: (2.1) PacBio HiFi software platform: PacBio WGS Variant Pipeline: https://github.com/PacificBiosciences/HiFi-human-WGS-WDL; PacBio HiFi tools: https://github.com/PacificBiosciences; (2.2) ONT software: MinKNOW with Guppy basecaller: https://community.nanoporetech.com/docs/analyse; ONT basecallers: https://github.com/nanoporetech; (2.3) Illumina software (srWGS): Illumina DRAGEN Bio-IT Platform (release 4.2): https://support.illumina.com/sequencing/sequencing_software/dragen-bio-it-platform.html.
References
Tsuji, S., Onodera, O., Goto, J., Nishizawa, M. & Study Group on Ataxic Diseases. Sporadic ataxias in Japan—a population-based epidemiological study. Cerebellum 7, 189–197 (2008).
Sullivan, R., Yau, W. Y., O’Connor, E. & Houlden, H. Spinocerebellar ataxia: an update. J. Neurol. 266, 533–544 (2019).
Flanigan, K. et al. Autosomal dominant spinocerebellar ataxia with sensory axonal neuropathy (SCA4): clinical description and genetic localization to chromosome 16q22.1. Am. J. Hum. Genet. 59, 392–399 (1996).
Vollger, M. R. et al. Segmental duplications and their variation in a complete human genome. Science 376, eabj6965 (2022).
Weisschuh, N. et al. Diagnostic genome sequencing improves diagnostic yield: a prospective single-centre study in 1000 patients with inherited eye diseases. J. Med. Genet. 61, 186–195 (2024).
Dolzhenko, E. et al. Resolving the unsolved: comprehensive assessment of tandem repeats at scale. Preprint at bioRxiv https://doi.org/10.1101/2023.05.12.540470 (2023).
Wallenius, J. et al. Exonic trinucleotide repeat expansions in ZFHX3 cause spinocerebellar ataxia type 4: a poly-glycine disease. Am. J. Hum. Genet. 111, 82–95 (2024).
Paucar, M. et al. Spinocerebellar ataxia type 4 is caused by a GGC expansion in the ZFHX3 gene and is associated with prominent dysautonomia and motor neuron signs. Preprint at medRxiv https://doi.org/10.1101/2023.10.03.23296230 (2023).
Chen, Z. et al. Adaptive long-read sequencing reveals GGC repeat expansion in ZFHX3 associated with spinocerebellar ataxia type 4. Mov. Disord. 39, 486–497 (2024).
Del Rocío Pérez Baca, M. et al. A novel neurodevelopmental syndrome caused by loss-of-function of the zinc finger homeobox 3 (ZFHX3) gene. Preprint at medRxiv https://doi.org/10.1101/2023.05.22.23289895 (2023).
Sagner, A. et al. A shared transcriptional code orchestrates temporal patterning of the central nervous system. PLoS Biol. 19, e3001450 (2021).
Ishiura, H., Tsuji, S. & Toda, T. Recent advances in CGG repeat diseases and a proposal of fragile X-associated tremor/ataxia syndrome, neuronal intranuclear inclusion disease, and oculophryngodistal myopathy (FNOP) spectrum disorder. J. Hum. Genet. 68, 169–174 (2023).
Boivin, M. & Charlet-Berguerand, N. Trinucleotide CGG repeat diseases: an expanding field of polyglycine proteins? Front. Genet. 13, 843014 (2022).
Liufu, T. et al. The polyG diseases: a new disease entity. Acta Neuropathol. Commun. 10, 79 (2022).
Rafehi, H. et al. An intronic GAA repeat expansion in FGF14 causes the autosomal-dominant adult-onset ataxia SCA27B/ATX-FGF14. Am. J. Hum. Genet. 110, 1018 (2023).
Pellerin, D. et al. Deep intronic FGF14 GAA repeat expansion in late-onset cerebellar ataxia. N. Engl. J. Med. 388, 128–141 (2023).
Famula, J. L. et al. Presence of middle cerebellar peduncle sign in FMR1 premutation carriers without tremor and ataxia. Front. Neurol. 9, 695 (2018).
Podar, I. V. et al. First case of adult onset neuronal intranuclear inclusion disease with both typical radiological signs and NOTCH2NLC repeat expansions in a Caucasian individual. Eur. J. Neurol. 30, 2854–2858 (2023).
Liu, M. et al. A comprehensive study of clinicopathological and genetic features of neuronal intranuclear inclusion disease. Neurol. Sci. 44, 3545–3556 (2023).
Hellenbroich, Y. et al. Spinocerebellar ataxia type 4 (SCA4): initial pathoanatomical study reveals widespread cerebellar and brainstem degeneration. J. Neural Transm. 113, 829–843 (2006).
Paul, S., Dansithong, W., Figueroa, K. P., Scoles, D. R. & Pulst, S. M. Staufen1 links RNA stress granules and autophagy in a model of neurodegeneration. Nat. Commun. 9, 3648 (2018).
Paul, S. et al. Staufen1 in human neurodegeneration. Ann. Neurol. 89, 1114–1128 (2021).
Paul, S. et al. Staufen impairs autophagy in neurodegeneration. Ann. Neurol. 93, 398–416 (2023).
Scoles, D. R. & Pulst, S. M. Spinocerebellar ataxia type 2. Adv. Exp. Med. Biol. 1049, 175–195 (2018).
Huynh, D. P., Figueroa, K., Hoang, N. & Pulst, S. M. Nuclear localization or inclusion body formation of ataxin-2 are not necessary for SCA2 pathogenesis in mouse or human. Nat. Genet. 26, 44–50 (2000).
Mizushima, N. & Yoshimori, T. How to interpret LC3 immunoblotting. Autophagy 3, 542–545 (2007).
Hill, A. C. & Hall, J. The MOE modification of RNA: origins and widescale impact on the oligonucleotide therapeutics field. Helv. Chim. Acta 106, e202200169 (2023).
Becker, L. A. et al. Therapeutic reduction of ataxin-2 extends lifespan and reduces pathology in TDP-43 mice. Nature 544, 367–371 (2017).
Scoles, D. R. et al. Antisense oligonucleotide therapy for spinocerebellar ataxia type 2. Nature 544, 362–366 (2017).
Rodriguez, C. M. & Todd, P. K. New pathologic mechanisms in nucleotide repeat expansion disorders. Neurobiol. Dis. 130, 104515 (2019).
Krans, A., Skariah, G., Zhang, Y., Bayly, B. & Todd, P. K. Neuropathology of RAN translation proteins in fragile X-associated tremor/ataxia syndrome. Acta Neuropathol. Commun. 7, 152 (2019).
Sellier, C. et al. Translation of expanded CGG repeats into FMRpolyG is pathogenic and may contribute to fragile X tremor ataxia syndrome. Neuron 93, 331–347 (2017).
Zhong, S. et al. Upstream open reading frame with NOTCH2NLC GGC expansion generates polyglycine aggregates and disrupts nucleocytoplasmic transport: implications for polyglycine diseases. Acta Neuropathol. 142, 1003–1023 (2021).
Boivin, M. et al. Translation of GGC repeat expansions into a toxic polyglycine protein in NIID defines a novel class of human genetic disorders: the polyG diseases. Neuron 109, 1825–1835 (2021).
Sone, J. et al. Long-read sequencing identifies GGC repeat expansions in NOTCH2NLC associated with neuronal intranuclear inclusion disease. Nat. Genet. 51, 1215–1221 (2019).
Ishiura, H. et al. Noncoding CGG repeat expansions in neuronal intranuclear inclusion disease, oculopharyngodistal myopathy and an overlapping disease. Nat. Genet. 51, 1222–1232 (2019).
Tian, Y. et al. Expansion of human-specific GGC repeat in neuronal intranuclear inclusion disease-related disorders. Am. J. Hum. Genet. 105, 166–176 (2019).
Karczewski, K. J. et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581, 434–443 (2020).
Cheng, H., Concepcion, G. T., Feng, X., Zhang, H. & Li, H. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat. Methods 18, 170–175 (2021).
Ranallo-Benavidez, T. R., Jaron, K. S. & Schatz, M. C. GenomeScope 2.0 and Smudgeplot for reference-free profiling of polyploid genomes. Nat. Commun. 11, 1432 (2020).
Kolmogorov, M. et al. Scalable Nanopore sequencing of human genomes provides a comprehensive view of haplotype-resolved variation and methylation. Nat. Methods 20, 1483–1492 (2023).
Poplin, R. et al. A universal SNP and small-indel variant caller using deep neural networks. Nat. Biotechnol. 36, 983–987 (2018).
Jeffares, D. C. et al. Transient structural variations have strong effects on quantitative traits and reproductive isolation in fission yeast. Nat. Commun. 8, 14061 (2017).
Kolmogorov, M., Yuan, J., Lin, Y. & Pevzner, P. A. Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 37, 540–546 (2019).
Smolka, M. et al. Detection of mosaic and population-level structural variants with Sniffles2. Nat. Biotechnol. https://doi.org/10.1038/s41587-023-02024-y (2024).
Li, H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100 (2018).
Shafin, K. et al. Haplotype-aware variant calling with PEPPER-Margin-DeepVariant enables high accuracy in nanopore long-reads. Nat. Methods 18, 1322–1332 (2021).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Liao, Y., Smyth, G. K. & Shi, W. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res. 47, e47 (2019).
Robinson, J. T. et al. Integrative Genomics Viewer. Nat. Biotechnol. 29, 24–26 (2011).
Dolzhenko, E. et al. ExpansionHunter: a sequence-graph-based tool to analyze variation in short tandem repeat regions. Bioinformatics 35, 4754–4756 (2019).
Takahashi, K. et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872 (2007).
Takahashi, K., Okita, K., Nakagawa, M. & Yamanaka, S. Induction of pluripotent stem cells from fibroblast cultures. Nat. Protoc. 2, 3081–3089 (2007).
Okita, K. et al. A more efficient method to generate integration-free human iPS cells. Nat. Methods 8, 409–412 (2011).
Fu, C. et al. The transcription factor ZFHX3 is crucial for the angiogenic function of hypoxia-inducible factor 1α in liver cancer cells. J. Biol. Chem. 295, 7060–7074 (2020).
Dong, X. Y. et al. ATBF1 inhibits estrogen receptor (ER) function by selectively competing with AIB1 for binding to the ER in ER-positive breast cancer cells. J. Biol. Chem. 285, 32801–32809 (2010).
Sturm, M. megSAP 2022_08. Zenodo https://doi.org/10.5281/zenodo.10817663 (2022).
Groß, C. & Admard, J. megLR 1.0.0. Zenodo https://doi.org/10.5281/zenodo.10820153 (2023).
Groß, C. expander. Zenodo https://doi.org/10.5281/zenodo.10820172 (2021).
Acknowledgements
We express our gratitude to the individuals with SCA4 and their relatives and caregivers. We thank L. Huynh, J. Hübener-Schmid, Q. Mao and the University of Utah Cell Imaging Core for their contributions. This work was supported by NIH grant R35 NS127253 to S.M.P. S.M.P. was partially supported by the Rindlisbacher Endowment for Research in Neurodegeneration. O.R. was partially supported by the Solve-RD project (European Union’s Horizon 2020 547 research and innovation program under grant agreement no. 779257). S.O. received funding from the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation; OS 647/1-1). S.Z. was supported by the DFG, grant 465281508. T.B.H. was supported by the DFG, grants 418081722 and 433158657. J. Park was supported by the Else Kröner-Fresenius-Stiftung Clinician Scientist program ‘PRECISE.net’. NGS methods were performed with the support of the DFG-funded NGS Competence Center Tübingen (INST 37/1049-1). We thank our colleagues from the Clinical Long-read Genome Initiative (lonGER consortium) for valuable discussion and exchange on methodological aspects of ONT LR-GS.
Author information
Authors and Affiliations
Contributions
K.P.F., M. Spielmann, O.R., S.O., T.B.H. and S.M.P. jointly supervised the research. K.P.F., N.C. and S.M.P. conceptualized and designed the experiments. K.P.F., C.G., E.B.-A., S.P., M.G., T.B.H., N.K., J. Park, K.H., C.D. and A.K. performed the experiments. K.P.F. performed real-time PCR analyses. S.P. performed western blotting. M.G. produced iPS cells and iPS cell-derived neurons. M.G. and A.K. performed immunohistological stains. K.P.F., S.P., M.G., J. Park, D.R.S., S.O. and T.B.H. performed statistical tests. K.P.F., E.B.-A., S.P., M.G., N.K., M. Sturm, N.C., J.A., Y.H., J. Pozojevic, S.B., T.F. and S.O. performed data analyses. K.P.F., N.K., C.Z., Y.H., S.Z., D.T., F.E., L.H., M.Z., T.K., O.R. and S.M.P. contributed patient materials or patient data. K.P.F., E.B.-A., N.K., M. Sturm, N.C., J. Park, D.R.S., A.K., M. Spielmann, O.R., S.O., T.B.H. and S.M.P. contributed to writing the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Genetics thanks Jianwen Deng, Clevio Nobrega and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–5 and Tables 1 and 2.
Source data
Source Data Fig. 2
Quantifications of histological data presented in Fig. 2.
Source Data Fig. 3
Full unprocessed blots associated with Fig. 3.
Source Data Fig. 3
Quantifications of blots presented in Fig. 3.
Source Data Fig. 4
Full unprocessed blots associated with Fig. 4.
Source Data Fig. 4
Quantifications of blots presented in Fig. 4.
Source Data Fig. 5
Full unprocessed blots associated with Fig. 5.
Source Data Fig. 5
Quantifications of blots presented in Fig. 5.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Figueroa, K.P., Gross, C., Buena-Atienza, E. et al. A GGC-repeat expansion in ZFHX3 encoding polyglycine causes spinocerebellar ataxia type 4 and impairs autophagy. Nat Genet (2024). https://doi.org/10.1038/s41588-024-01719-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41588-024-01719-5