Abstract
To examine the transmission dynamics of Mycobacterium tuberculosis (Mtb) isolated from tuberculosis patients in Ho Chi Minh City, Vietnam, we sequenced the whole genomes of 1,635 isolates and compared these with 3,144 isolates from elsewhere. The data identify an underlying burden of disease caused by the endemic Mtb lineage 1 associated with the activation of long-term latent infection, and a threefold higher burden associated with the more recently introduced Beijing lineage and lineage 4 Mtb strains. We find that Beijing lineage Mtb is frequently transferred between Vietnam and other countries, and detect higher levels of transmission of Beijing lineage strains within this host population than the endemic lineage 1 Mtb. Screening for parallel evolution of Beijing lineage-associated SNPs in other Mtb lineages as a signal of positive selection, we identify an alteration in the ESX-5 type VII-secreted protein EsxW, which could potentially contribute to the enhanced transmission of Beijing lineage Mtb in Vietnamese and other host populations.
Similar content being viewed by others
Main
Tuberculosis (TB) is a leading cause of death from infectious disease and the global burden is now higher than at any point in history1,2. Despite coordinated efforts to control TB transmission, the factors contributing to its successful spread remain poorly understood. Vietnam is identified as 1 of 30 high-burden countries for TB and multi-drug-resistant TB with an incidence of 137 TB cases per 100,000 individuals in 20152. Recent phylogenomic analyses of the causative agent Mycobacterium tuberculosis (Mtb) in other high-prevalence regions have provided insights into the complex processes underlying TB transmission3,4,5.
Results
Genetic diversity and drug resistance
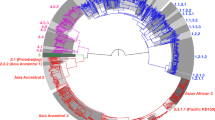
To characterize the diversity of Mtb circulating in Ho Chi Minh City (HCMC), we sequenced the genomes of 1,635 isolates (Supplementary Table 1) obtained from 2,091 human immunodeficiency virus (HIV)-negative, smear-positive adults (≥18 years old) commencing anti-TB therapy at district TB units (DTUs) in eight districts of HCMC between December 2008 and July 2011 (see Methods). This identified 73,718 SNPs, which we used to reconstruct a maximum-likelihood phylogeny (Fig. 1a) and to assign lineages6. The majority of isolates (n = 957, 59%) belonged to lineage 2.2.1, a subgroup of the Beijing lineage (2.2). Lineage 1 (Indo-Oceanic lineage; n = 388, 23.7%) and lineage 4 (Euro-American lineage; n = 192, 11.7%) were also common. A single isolate belonged to lineage 3 (East African-Indian lineage) and was excluded from further analysis. The distribution of the lineages did not change during the 2.5-year period of study (Fig. 1b), and was in agreement with previous genotyping studies in urban areas of Vietnam (≥50% 2.2/Beijing lineage and ~20% lineage 1.1/EIA in Hanoi and HCMC, 1998–2009)7,8,9,10,11. Known antimicrobial resistance mutations were detected in all lineages but were more frequent in Beijing sublineage 2.2.1 (Table 1), consistent with earlier reports from Vietnam7,8,9,11. In particular, Beijing sublineage 2.2.1 was enriched for mutations associated with resistance to streptomycin (odds ratio (OR) 4.6 [95% confidence interval (CI) 3.6–6.0], P = 1 × 10−15), isoniazid (OR 1.7 [1.3–2.1], P = 3 × 10−5), rifampicin (OR 5.4 [2.5–13.2], P = 1 × 10−7) and ethambutol (OR 5.6 [2.6–13.7], P = 1 × 10−7), using Fisher’s exact test to compare it with all non-2.2.1 Mtb isolates.
a, Maximum-likelihood phylogeny of 1,635 Mtb isolates collected from TB patients in HCMC, with backgrounds shaded by lineage. The exterior rings indicate the presence of known antimicrobial resistance-associated mutations (colored by drug, according to the legend in the top right). b, Frequency distribution of lineages by month. c, Frequency distribution of lineages by patient age group.
Mtb lineage and host demographics
While the majority of TB patients were male (74%, typical for TB studies in Vietnam and elsewhere8,9,10,12), the Beijing sublineage was significantly associated with TB in females (OR 1.28 [95% CI 1.01–1.62], P = 0.043 using Fisher’s exact test; Table 1), consistent with previous observations in Vietnam8 and Nepal13. Beijing sublineage 2.2.1 was also significantly associated with younger people: its frequency declined with age, from 74% of cases in <20 year olds to 50% in ≥60 year olds (P = 0.0023 Fisher’s exact test, P = 0.0024 linear trend test; Fig. 1c). In contrast, lineage 1 was significantly associated with males (25% of male cases versus 19% of females, P = 0.017) and increased with age regardless of gender, from 12% in <20 year olds to 35% in ≥60 year olds (P = 0.0007 Fisher’s exact test, P = 0.0014 linear trend test; Fig. 1c). These data confirm that Beijing sublineage 2.2.1 is capable of causing active disease in a wider demographic range of the Vietnamese host population, particularly among females and younger people, than the endemic lineage 17–10, which is associated with the more typical profile of TB susceptibility that is skewed towards males (the M/F prevalence ratio for smear-positive TB was recently estimated at 2.5 [95% CI 2.07–3.04], based on 40 surveys in 22 countries12) and older people2,14.
Mtb lineages display distinct local transmission dynamics
We hypothesized that the Beijing lineage or sublineage 2.2.1 was more transmissible than lineage 1, and/or more capable of causing active disease in infected hosts, in the local Vietnamese Kinh host population. To investigate this, we used the whole genome phylogeny to compare diversity metrics for each lineage (Fig. 2). Terminal branch lengths, which represent an upper bound of evolutionary time since transmission for each Mtb case, were significantly shorter for Beijing sublineage 2.2.1 Mtb isolates (median 8 SNPs) than for non-Beijing lineage isolates (lineage 1: median 53 SNPs, P < 1 × 10−15 using the Kolmogorov–Smirnov test; lineage 2.1: 30 SNPs, P < 1 × 10−6; lineage 4: 17 SNPs, P < 1 × 10−9), and slightly shorter than Beijing sublineage 2.2.2 isolates (9 SNPs, P = 0.02) (Fig. 2a). The distribution of mean node-to-tip distances for all internal nodes was skewed significantly lower within the Beijing sublineage 2.2.1 compared with the rest of the tree (median 16 SNPs compared with 62, 57, 39 and 60 SNPs for lineages 1, 2.1, 2.2.2 and 4, respectively; P < 0.0015 in all cases).
a, Distributions of terminal branch lengths for the 1,635-strain phylogeny. b, Proportion of isolates from each lineage (colored as in panel a) that belong to clusters (y axis) defined at different thresholds for maximum pairwise SNP distances (x axis). c, Stacked area plot showing the number of clusters (left y axis) within each lineage (colored as in a) identified using different maximum patristic distance thresholds to define clusters (x axis). Black points indicate the proportion of isolates in clusters (right y axis) defined at the different patrisitic distance thresholds (x axis).
To better understand the differences in the transmission dynamics, we explored the distribution of potential transmission clusters using a range of maximum pairwise SNP distance thresholds to define a cluster15 (Fig. 2b,c). Using the smallest cutoff of 5 SNPs (transmission age of <5 years), there were n = 109 clusters, of which 76 (70%) belonged to Beijing sublineage 2.2.1; these had a mean size of 2.4 strains per cluster, compared with a mean of 2.1 for other clusters (Fig. 2b,c). Using cutoffs of 10 and 20 SNPs, there were n = 164 and n = 220 clusters, respectively; of these, 118 (72%) and 156 (71%), respectively, were Beijing 2.2.1 and these showed significantly larger cluster sizes (means, 2.5 and 3.2 strains per cluster) than those of other lineages (means, 2.2 and 2.4). Notably, the proportion of cases that belonged to transmission clusters was significantly lower among lineage 1 cases (7.7% at the 10 SNP threshold) compared with lineage 4 cases (20.3%, P = 2 × 10−5), which in turn was significantly lower than among Beijing lineage cases (31.5%, P = 3 × 10−3; see Fig. 2b). Therefore, sublineage 2.2.1 infections were more likely to result from recent detectable transmission within the local population, despite our study having low power to detect recent transmission due to sampling only a small fraction of all incident cases in HCMC (~30% of those in the study districts, <10% across HCMC, and restricted to only new cases of smear-positive, culture-positive HIV-negative TB; see Methods). Household data were not available; however, pairs of TB cases whose infecting Mtb strains were separated by ≤10 or ≤20 SNPs were significantly more likely to be diagnosed in the same DTU than more distantly related pairs of the same lineage (27% or 21%, versus 15% among strains separated by >20 SNPs; P < 1 × 10−12, see Supplementary Fig. 1). This phenomenon was observed for both Beijing 2.2.1 and non-Beijing clusters, but the effect was significantly less for the Beijing lineage (of strain pairs separated by <20 SNPs, 21% of Beijing pairs versus 29% of non-Beijing pairs were diagnosed in the same DTUs; P = 0.036, see Supplementary Fig. 1), suggesting that they may be more readily transmitted across greater geographical distances within HCMC than other Mtb lineages. This could potentially be associated with the higher frequency of Beijing lineage TB among younger adults (Fig. 1c), who may be more likely to travel regularly between districts, promoting onward transmission; however, the age distribution among transmission clusters did not differ by lineage. Taken together, the phylogenomic data identify significant differences in the transmission dynamics between the various Mtb lineages circulating in HCMC, and suggest that newly diagnosed cases of lineage 1 Mtb in this population often result from activation of longer-term latent infections with many private SNPs and no evidence of recent transmission, while new cases of Beijing sublineage 2.2.1 Mtb often result from more recent transmission and a shorter time to develop active disease.
Geographical relationships of Vietnamese Mtb strains
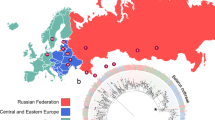
It has been suggested that the Beijing lineage is slowly displacing the resident lineage 1 strains in Vietnam, following the introduction of the Beijing strain into urban areas and subsequent spread to rural areas where lineage 1 still dominates8,10. Our data are consistent with this, showing a higher frequency of the Beijing lineage (65%) among Mtb infections in HCMC in 2008–2011 compared with the frequencies reported in the city a decade earlier7 (53% in 1998), or in rural areas outside the city8 (32–37% in 2003–2005). We therefore hypothesized that Beijing 2.2.1 isolates from HCMC may represent a locally established epidemic subclade of the Beijing lineage, similar to that previously described in Russia3. To investigate this, we combined our HCMC Mtb genome data with 3,146 publicly available Mtb whole-genome sequences from Russia3, Malawi4,5, Argentina16 and China17; and globally dispersed lineage 1 and 2 genomes18,19,20 (Supplementary Table 2); then inferred phylogenies for each lineage (Fig. 3). HCMC lineage 1 strains were quite distinct from those in other locations (Fig. 3a), with little evidence of transfer between Vietnam and other regions. The vast majority (n = 319, 82%) belonged to a localized subclade (1.1.1.16) that included only seven previously sequenced strains, all of which were from Vietnam. A further 46 (12%) of the HCMC strains belonged to a related sister clade (1.1.1) that also included strains from neighbouring Cambodia (n = 1) and Thailand (n = 2); the remainder (n = 23, 6%) belonged to subclade 1.2.1, in which the HCMC strains were intermingled with others from the Philippines (n = 9) and China (n = 1). These data suggest that lineage 1-associated TB in HCMC results mainly from a local endemic Mtb population. In contrast, Beijing 2.2.1 isolates from HCMC formed several distinct clusters that each shared a recent common ancestor with isolates from outside Vietnam (Fig. 3b). Notably, isolates from Russia, Malawi, China and numerous other countries were interspersed throughout the HCMC Beijing 2.2.1 population (Fig. 3b), suggesting multiple, frequent transfers between host populations in HCMC and other geographic regions. HCMC lineage 4 isolates were drawn from eight of the ten recognized sublineages6 (including those identified as specialist, generalist and intermediate in their geographic range21) and were interspersed with isolates from other geographical locations, consistent with multiple imports into HCMC (Fig. 3c). In further support of these observations, stochastic mapping of locations onto the phylogenies predicted dozens of strain transfer events between Vietnam and other locations for lineages 2 and 4, but not for lineage 1 (Fig. 3d), strongly supporting that Mtb sublineage 1.1.1.1 is endemic in Vietnam.
HCMC isolates are colored gray, isolates from four other localized studies are colored as in d and other locations are shown in black. a, Lineage 1 (n = 675 genomes). b, Lineage 2 (n = 1,871 genomes). c, Lineage 4 (n = 2,066 genomes). d, Number of transfers between Vietnam and other locations predicted by stochastic mapping of locations onto the lineage 2 and 4 trees.
Beijing lineage-defining SNPs under positive selection
The population structure (Figs. 1 and 2) provides evidence that Beijing lineage strains are more transmissible within this HIV-negative HCMC population than are other Mtb lineages. Genomic evidence for enhanced transmission of the Beijing lineage has been documented in Russia (associated with antimicrobial resistance)3 and Malawi (independent of antimicrobial resistance)4. While antimicrobial resistance was common among HCMC Beijing lineage isolates, the majority of transmission clusters (defined by ≤10 SNPs) comprised groups of isolates that did not share any known resistance mutations that could account for their transmission success (Supplementary Fig. 2). This is consistent with previous reports that the Beijing lineage is highly transmissible and more likely to progress to active disease in various host populations and is also more virulent and less pro-inflammatory in various cellular assays, independent of antimicrobial resistance22,23,24,25. We therefore aimed to interrogate the Mtb genome data to identify mutations that may contribute to the success of the Beijing lineage (2.2). Evolutionary convergence has previously been used as a signal of positive selection to identify mutations associated with antimicrobial resistance in Mtb26,27. We reasoned that advantageous polymorphisms contributing to the enhanced transmissibility of lineage 2.2 should be fixed in this lineage, and should also be under positive selection that is detectable as convergent or parallel evolution at the same variant sites in other lineages. We identified a total of 424 homoplasic non-synonymous SNPs (nsSNPs) across the HCMC phylogeny. The most frequent of these occurred in genes in which convergent evolution has previously been associated with antimicrobial resistance including gidB, embB, gyrA, rpo and inhA26, which together accounted for 12.4% of all homoplasic nsSNPs. The distribution of common homoplasic nsSNPs in these genes is shown in Supplementary Fig. 3; in particular, rifampicin resistance-associated mutations in rpoB and ethambutol-associated mutations in embB arose independently many times in Beijing sublineage 2.2.1 (n = 33/35 rpoB-450 mutations; n = 16/17 embB-306 mutations; n = 8/10 embB-406 mutations). The homoplasic nsSNPs included three that arose on the branch defining lineage 2.2 and also elsewhere in the HCMC tree (Table 2, Supplementary Fig. 4). One was a mutation in esxW (Rv3620c) codon 2 (EsxW-Thr2Ala), which arose on nine other branches (6 times in lineage 4, 3 times in lineage 1; see Supplementary Fig. 4) and showed evidence of onward transmission on 4/9 occasions. Comparison with the global tree detected the same esxW mutation on a further 10 lineage 4 branches in Malawi and Russia, with onward transmission detected on 6/10 occasions. The other two mutations were in Rv3081 (conserved hypothetical protein) and gidB (mutations in which are often associated with streptomycin resistance) and arose less frequently (Table 2). In contrast, homoplasic nsSNPs on the branches defining lineage 1 or 4 were each detected on only 1–2 other branches in the HCMC tree and no additional branches of the global tree (Supplementary Table 3). No homoplasic SNPs were associated with sublineage 2.2.1, and although synonymous or intergenic SNPs can have functional consequences, we found no such homoplasies associated with the Beijing or other lineages.
EsxW alteration
EsxW is included in multiple Mtb vaccines currently under development (including H65, ID83, ID93)28,29,30,31 due to its demonstrated immunogenicity in mice, safety and immunogenicity in non-human primates31, demonstrated T-cell targeting in humans32 and epitopes predicted to bind a wide range of human HLA-DRB1 alleles28,33. Hence, we considered whether the EsxW-Thr2Ala alteration could affect epitope binding. However, residue 2 lies in the amino-terminal loop of EsxW (Fig. 4e), outside the experimentally demonstrated epitope region (residues 24–34)34. In silico epitope binding analysis on the Beijing and non-Beijing EsxW protein sequences using the Immune Epitope Database Analysis Resource35 predicted HLA binding to the first 9–10 residues of EsxW, but no differences in binding affinities for the wild-type and Thr2Ala mutant alleles. This is consistent with experimental data showing that immunization with ID93 (a recombinant fusion protein containing H37Rv (lineage 4) wild-type alleles of EsxW/V) is protective against Beijing (2Ala) as well as non-Beijing (2Thr) strains30,31.
a, Variation in the promoter region of esxW and other CFP10 paralogs extracted from H37Rv (lineage 4). The sites are colored by conservation; the coordinates are relative to the start codon. b, Variation in the protein-coding region. The tree shows the maximum-likelihood phylogeny inferred from amino-acid sequences; the box indicates QILSS proteins. The alignment of the protein-coding DNA sequence is shown, colored by conservation; the arrow indicates the site of the homoplasic SNP in esxW (A to G) resulting in Thr to Ala substitution at EsxW protein residue 2. c,d, RNA-Seq results for esxW and QILSS/ESX-5 paralogs measured in four Mtb isolate pairs, each including a lineage 1 or 4 EsxW-2Ala mutant and its genetically closest EsxW-2Thr relative, following 24 h macrophage infection. mRNA levels were estimated from read counts uniquely mapping to the region −21 to +9 for each gene; normalized to total reads uniquely mapping to the locus encoding the ESX-5 machinery (and esxM) in each isolate. The boxes indicate the interquartile range and the black lines indicate the median. d, Difference between the 2Ala mutant and the 2Thr wild-type for each pair, relative to the wild-type expression level. e, Structural model of EsxW–EsxV heterodimer. EsxV is shown as a surface (gray) and EsxW as a ribbon (red) with key residues shown as labeled sticks. f, Comparison of biophysical measurements of heterodimer binding affinity between wild-type and mutant EsxW. All binding curves were determined across four replicates by microscale thermophoresis, and are represented as the mean ± standard deviation.
Next we considered whether the EsxW-Thr2Ala alteration could affect gene expression or protein structure and function. esxW is 1 of 23 esx genes in the Mtb genome, including 11 clustered pairs of esx genes whose products form heterodimers that are each secreted by 1 of 5 type VII secretion systems (T7SS; ESX-1 to ESX-5). The most-studied of these pairs is esxB (CFP10)/esxA (ESAT-6), secreted by ESX-1 and encoded in the RD1 locus, which also encodes the ESX-1 system. EsxW and its heterodimerization partner EsxV are encoded by adjacent genes in the RD8 locus and secreted by ESX-5. The ESX-5 system is the most recently evolved T7SS in Mtb and is present only in the slow-growing Mycobacteria36,37 (including the Mtb complex, M. leprae, M. ulcerans and M. marinum). ESX-5 is unique among the Mtb T7SS in that it secretes most of the PE/PPE proteins38, which are also unique to slow-growing Mycobacteria, comprise a substantial amount of protein-coding capacity (~10%) in the Mtb genome and play various roles in virulence39,40; and it is associated with five pairs of Esx proteins, resulting from duplication and expansion of esxM and esxN (which are encoded within the locus encoding the ESX-5 T7SS machinery) to create four paralogous copies elsewhere in the Mtb genome41 (Fig. 4). Each pair includes a member of the QILSS family (EsxM paralog, including EsxW) and a member of the Mtb9.9 family (EsxN paralog, including EsxV), which partner to form heterodimers. The paralogous proteins differ from one another by just a few amino acids (Fig. 4b), and the reason for this apparent redundancy is not clear. One pair, EsxJ/EsxI, has been shown to play a role in substrate selection for ESX-5 secretion42, and it is hypothesized that the other paralogs including EsxW/V play a similar role41. Notably, two of the loci encoding QILSS paralogs (EsxP/O, RD5; and EsxW/V, RD8) are missing from the M. bovis BCG vaccine strain.
The upstream sequences of esxW and its homologs differ substantially (Fig. 4a). This suggests that their expression is subject to different regulatory controls, which could provide a reason for their expansion in the Mtb genome despite the lack of differentiation at the protein level43 (Fig. 4b); indeed, it has been shown that the different QILSS family members are all expressed at different levels by the Mtb strain H37Rv during growth in broth, sputum and macrophages44. This uniqueness also allows unambiguous read mapping and confident SNP calling at the N-terminal region of EsxW; however, we also used PCR and capillary sequencing to confirm the EsxW-2Ala allele in all non-lineage 2 strains in which the SNP was identified from Illumina reads (see Methods). We sought to investigate the expression of QILSS family proteins, and the potential impact of the EsxW-Thr2Ala alteration on gene expression, during growth in macrophages. For these experiments, we selected four non-Beijing Mtb isolates harbouring the EsxW-2Ala allele, and the closest genetic relative of each with the wild-type allele EsxW-2Thr, and used RNA sequencing (RNA-seq) to measure genome-wide Mtb expression levels (see Methods). The results showed that the ESX-5 system was highly expressed in all isolates following 24 h of growth in macrophages (mean 0.25% of total Mtb RNA-Seq reads); however, the QILSS paralogs were expressed at different levels relative to ESX-5 (Fig. 4c). Compared with esxM, which is encoded within the ESX-5 locus itself and was expressed at the highest levels in all isolates, esxK and esxP were expressed at intermediate levels (mean 42% of esxM level) and esxJ and esxW were expressed at low levels (mean 2.2% of esxM level). There were no significant differences between the EsxW-2Ala mutant versus the wild-type in terms of Mtb growth in macrophages (measured at 4, 7 or 11 days post-infection), or in esxW (Fig. 4d), ESX-5 or global gene expression measured at 24 h post-infection. It has been reported that esxW was significantly expressed in the lungs of Mtb aerosol-infected mice45; hence, future experiments with wild-type and mutant EsxW in this animal system could potentially help to unravel its functional effects.
Given the proposed role for the EsxW/V heterodimer in substrate selection for ESX-5 secretion41, we investigated whether the EsxW-Thr2Ala alteration could affect protein structure and function. In M. canetti, the reference Mtb genome H37Rv and the majority of non-Beijing Mtb isolates, EsxW carries the polar threonine (codon ACC) at residue 2, while the other QILSS proteins in the Mtb complex and other slow-growing Mycobacteria carry the hydrophobic alanine (GCC) at this position (Fig. 4b). In the Mtb Beijing lineage, EsxW residue 2 is converted to the more typical residue alanine (GCC), making it identical at the protein level to EsxJ, which has been shown to be involved in substrate selection for ESX-5 secretion42. We analyzed the quantitative effects of the Thr2Ala alteration on the stability and affinity of the EsxW protomer and the EsxW/V heterodimer, using computational modeling and direct biophysical experiments (see Methods and Fig. 4e). This indicated that the 2Ala alteration was likely to lead to a mild increase in the affinity and stability of the heterodimer complex (mean ΔΔG 0.28 ± 0.06 kcal mol−1), and this was supported by biophysical measurements that showed the mutant bound slightly more strongly to EsxV than the wild-type (Kdwt = 0.6 µM; KdT2A = 0.4 µM; P = 0.05 using a two-tailed t-test; Fig. 4f). We hypothesize that, if EsxW/V does indeed play a role in substrate selection for ESX-5 secretion, then increased stability of the heterodimer and/or the hydrophobicity of EsxW-2Ala could potentially affect the efficiency of secretion of certain PE/PPE proteins. This could thereby have downstream impacts on one or more of the known functions of ESX-5 including inflammasome activation, IL-1β secretion or escape from macrophages46, any of which could potentially work to promote transmission between hosts.
Discussion
The shorter terminal branch lengths and node-to-tip distances for local Beijing lineage Mtb (Fig. 2) could be explained by: a slower mutation rate in the Beijing lineage, resulting in slower accumulation of SNPs over time; sampling bias, whereby new cases of active TB arising in the study population were more likely to be detected and included in the study if they were caused by Beijing lineage strains; or strain-specific transmission dynamics in the study population, whereby the average time taken to progress to active disease is shorter for the Beijing lineage than for other Mtb strains. The mutation rate for Mtb has been estimated at ~0.5 SNPs per year for lineage 4, and about two times faster for Beijing strains47 (lineage 1 has not been specifically measured but is assumed to be similar to lineage 4, which is considered typical). Hence, if all Mtb lineages were subject to the same transmission dynamics within the study area, we would expect to see longer terminal branch lengths for Beijing lineage isolates, whereas here we observe the opposite (significantly shorter branch lengths). Patients were recruited into the study following positive diagnosis at eight central DTUs in HCMC (Supplementary Fig. 5). The identity of the infecting Mtb lineages was not known at the time of recruitment, and we are unaware of any factors that would bias the inclusion of Beijing lineage cases over others presenting to the clinics for treatment; hence, we conclude that the frequency of infections with Beijing strains, and their shorter branch lengths, reflect differences in the transmission dynamics of these strains within the study population.
The lack of transfer of Mtb sublineage 1.1.1.1 between Vietnam and other geographical locations (Fig. 3) may be associated with adaptation to the local host population with which it has co-evolved for centuries, similar to the ‘host-specialist’ clades recently identified within lineage 421. In contrast, while the direction of transfer of lineage 2.2.1 strains between HCMC and other geographical regions cannot be determined from our data, the frequency of transfer events and the scale of diversity among the HCMC strains (Fig. 3) make it unlikely that the rise of Beijing sublineage 2.2.1 in HCMC represents clonal spread of a locally established subclade. Regardless of the direction, the frequency of transfer between Vietnam and diverse geographically dispersed populations supports previous contentions that the Beijing lineage is a host generalist, capable of moving between ethnically diverse host populations24.
While the mechanism remains to be elucidated, our results provide evidence that the Beijing lineage carries a variant of esxW that is under positive selection in natural Mtb populations. This is consistent with the idea that the protein is important for host interactions, potentially through substrate selection for ESX-5 secretion under certain conditions. Immunizing against EsxW has already been shown to be protective against infection29,30,31,32, and positive selection for this protein suggests that vaccines including it are likely to remain effective in the long term.
Taken together, our data show that the burden of TB in HCMC comprises: an underlying burden of disease caused by the endemic lineage 1 Mtb strains (24% of all TB cases), which disproportionately affect men and older people and are associated more with activation of long-term latent infection than short-term transmission clusters; and an additional disease burden caused by the more recently introduced lineage 2 and 4 Mtb strains (76% of all TB cases). In particular, Beijing sublineage 2.2.1 was associated with a wider demographic host range, infecting women and young people significantly more frequently than other lineages, and was associated with a shorter time to active disease and frequent onward local transmission. One-third of all Beijing strains were involved in transmission clusters (≤10 SNPs), and these were associated with wider geographic dispersal within HCMC. Notably, 75% of TB cases associated with transmission clusters involved the Beijing lineage, accounting for 20% of all cases included in the genomic study.
Importantly, these data show that not all Mtb strains are equal: genetically diverse strains display distinct transmission dynamics even within a single localized host population, suggesting that a more detailed understanding of lineage-specific variation in Mtb could be informative to tailor local TB control in HCMC and other settings. For example, TB contact tracing is commonly used in low-incidence high-income countries, but results vary in high-incidence low- and middle-income countries and there is a need to prioritize resources towards cases where contact tracing is most likely to yield results, which includes considering the likelihood of transmission and progression to active disease within the time frame of a contact tracing program (1–2 years)48,49. In HCMC, it may thus be advantageous to direct contact tracing resources towards Beijing lineage cases, as they pose the greatest risk of onward transmission resulting in new active TB cases.
Methods
Bacterial isolates used in this study
Between December 2008 and July 2011, 2,091 individuals of the Vietnamese Kinh ethnic group attending the outpatient department of Pham Ngoc Thach Hospital or from 8 DTUs (Districts 1, 4, 5, 6 and 8, Tan Binh, Binh Thanh and Phu Nhuan) in HCMC were recruited into a clinical study investigating predictors of failure and relapse in isoniazid-resistant TB50. The 8 TB units were chosen for inclusion from among the 24 servicing HCMC as they are centrally located and close to the TB reference laboratory, which was essential for the logistics of sample collection and processing (see the map in Supplementary Fig. 5). Inclusion criteria were: 18 years or older, negative HIV test, provision of written informed consent, smear-positive pulmonary TB. Exclusion criteria were: under 18 years of age, HIV infected, unable or unwilling to provide consent, pregnancy, prior history of TB antibiotic therapy, will receive TB-DOTS (directly observed treatment, short-course) outside the study centres. Over the 2.5-year study period, there were n = 5,036 new smear-positive cases of TB (HIV positive and negative) at the district tuberculosis units, of which n = 2,091 were eligible for recruitment. Of these, n = 1,822 Mtb strains were isolated from the study participants. The annual incidence of pulmonary TB in HCMC is ~82,000, of which ~11,000 are in HCMC; therefore, our sample represents ~6.6% of all cases in HCMC during the 2.5-year study period, and >30% of cases in the study districts.
Ethics
The study protocol was approved by the Institutional Research Board of Pham Ngoc Thach Hospital (the supervisory institution of the District TB Units in southern Vietnam), Ho Chi Minh City Health Services and the Oxford University Tropical Research Ethics Committee, UK (Oxtrec 030-07). Written informed consent was obtained from all patients.
DNA extraction and sequencing
Mtb isolates were subcultured on Lowenstein Jensen media and DNA was extracted at the Oxford University Clinical Research Unit in HCMC using the cetyl trimethylammonium bromide extraction protocol as described previously51. DNA was successfully obtained from n = 1,728 isolates and shipped to the University of Melbourne for whole-genome sequencing. DNA extracts were purified using the AxyPrep Mag PCR Normalizer Protocol before library preparation. A total of n = 1,655 DNA samples passed quality control, were included for sequencing and subjected to library preparation using the Nextera XT protocol. Libraries were quantified using Quant-iT PicoGreen (dsDNA kit, Invitrogen), then normalized and pooled to 4 nM concentration. DNA underwent 150-base pair (bp) paired-end sequencing (Rapid mode v2) on the Illumina HiSeq 2500 platform (Illumina, San Diego). Sequence data were excluded for n = 19 Mtb isolates that yielded less than the pre-established criteria of 10 times the mean read depth across the Mtb genome, as SNPs cannot be reliably called below this depth. Sequence data were successfully generated for n = 1,635 Mtb isolates from HCMC (representing 90% of those isolated from eligible patients in the cohort) with a median of three million reads per sample, providing a median of 99.2% coverage and 86 times the depth for each Mtb genome (Supplementary Table 1). To confirm the esxW codon 2 SNP, we performed PCR and capillary sequencing of the region flanking the SNP in all non-Beijing lineage strains in which the SNP was detected from Illumina data. The primers used are listed in Supplementary Table 4.
Mtb genome data
Mtb genome data generated from 1,635 isolates (Fig. 1) have been deposited in the NCBI BioProject database (accession ID: PRJNA355614; see the URLs section). A total of 3,144 Mtb genomes were included in the analysis in Fig. 3, comprising data from localized studies: 1,032 from Russia3, 1,621 from Malawi4,5, 248 Argentina16 and 78 from China17; as well as 106 globally dispersed lineage 2 genomes18 and 59 globally dispersed lineage 1 genomes19,20. Illumina Mtb genome sequences from various previously published studies were downloaded from the European Nucleotide Archive (see the URLs section, individual accession numbers are given in Supplementary Table 2). The H37Rv reference genome sequence (NC_000962.3) was used for all reference-driven analyses.
SNP analysis
Sequence reads were mapped to the H37Rv reference genome using the RedDog pipeline v0.5 (see the URLs section). Briefly, Bowtie2 v2.2.3 was used for read alignment with the sensitive-local algorithm and the maximum insert length set to 2,000 (via the -x parameter)52 and variant sites (that is, SNPs) were called using SAMTools v0.1.1953. SNPs located in previously reported repetitive regions of the genome were excluded before phylogenetic analysis54,55 (Supplementary Table 5); sites for which a definitive allele call could not be made in at least 99.5% of all isolate sequences were also excluded from the set of SNPs used for phylogenetic analysis. Two SNP alignments were compiled for analysis: one comprising the 1,635 HCMC isolates (total 73,718 SNPs), and one comprising all 4,779 isolates (including the HCMC isolates and the global collections downloaded from public data; total 133,492 SNPs).
In silico lineage and antimicrobial resistance typing
Mykrobe Predictor v0.3.6 was used to analyze raw Illumina reads generated from HCMC Mtb isolates and: assign each isolate to one of the seven Mtb lineages; and detect known resistance-associated polymorphisms56 (summarized in Table 1, individual mutation calls are provided in Supplementary Table 1). All Mtb isolates were further assigned to sublineages by comparing SNPs identified using RedDog with those used in the haplotyping scheme defined in ref. 6 (lineage assignments are in Supplementary Tables 1 and 2).
Phylogenomic analyses
Maximum-likelihood phylogenetic trees were inferred using RAxML v7.7.257 for: all HCMC isolates (presented in Fig. 1); and each of lineages 1, 2 and 4 using combined data from the HCMC isolates and available public data (presented in Fig. 3; see the isolates list in Supplementary Tables 1 and 2). The trees presented are those with the highest likelihood from five replicate runs, constructed using the general time reversible model of nucleotide substitution and a Gamma model of rate heterogeneity to analyze a concatenated alignment of SNP alleles. An approximate maximum-likelihood tree containing all data (HCMC isolates and available public data) was inferred using FastTree v2.1.858. Ancestral sequence reconstruction was performed for the HCMC tree and combined tree using FastML v3.1 to infer the sequence alignment at each internal node of the maximum-likelihood phylogeny59. Substitution events occurring on each branch of the tree were extracted by comparing the joint reconstruction sequences for the parent and child nodes; these data were used to identify homoplasic SNPs, and to identify lineage-specific polymorphisms as well as independent occurrences of those polymorphisms outside the lineage of interest (data in Table 2). Terminal branch lengths reported are the number of substitutions (SNPs) mapped to each terminal branch (data in Fig. 2a) and were compared to the Beijing 2.2.1 sublineage using two-sample Kolmogorov–Smirnov tests. Metrics for genetic diversity and tree topology were calculated from the phylogenies using R. Node-to-tip distances showed similar variances within groups (standard deviations of 27–75 SNPs) and all groups were compared to the Beijing 2.2.1 sublineage using two-sample Kolmogorov–Smirnov tests. Clusters were defined as subtrees for which the maximum patristic distance between descendant tips fell below a specified threshold (data in Fig. 2b,c). Each cluster was checked to determine whether all members of the cluster shared any of the antimicrobial resistance mutations identified by Mykrobe Predictor; clusters in which no known antimicrobial resistance mutation was conserved in all members of the cluster are reported as not explained by antimicrobial resistance (data in Supplementary Fig. 2).
Phylogeography analysis
Transmission between geographical regions was assessed separately for lineage 1, 2 and 4 trees using an implementation of stochastic mapping on phylogenies (SIMMAP) implemented in the phytools v0.5 package for R60,61. Region of origin was treated as a discrete trait and mapped to each tree using the ARD model (which allows each region-to-region transfer rate to vary independently) with 100 replicates. The results reported (Fig. 3d) are the median values for the number of transitions to Vietnam from any other region, summarized from 100 replicate mappings for each tree.
Esx sequence analysis
Esx protein sequences were extracted from the H37Rv reference genome using Artemis, aligned using Muscle and subjected to phylogenetic inference using PhyML v3.0 (tree in Fig. 4). DNA sequences flanking the start codon of each esx gene were extracted from the H37Rv reference genome using Artemis and aligned and visualized using JalView v2.6.1 (Fig. 4).
Macrophage infections
Mtb infection of macrophages (THP1 human cell line, 88081201, Sigma-Aldrich) has been described previously62. To study Mtb growth in macrophages, 2.5 × 105 THP1 cells were seeded in 24-well cell culture plates and infected with Mtb isolates in triplicate at a multiplicity of infection (MOI) of 1. At 4 h post-infection, extracellular bacteria were removed by washing. At 0, 4, 7 and 11 days post-infection, intracellular bacteria were harvested and plated on Middlebrook 7H10 agar plates. Colony-forming unit counts resulted after 3 to 4 weeks of incubation at 37 °C. To measure genome-wide Mtb expression levels, confluent monolayers of 1.5 × 107 THP1 in 75 cm2 cell culture flasks were infected with Mtb isolates at a MOI 4. At 4 h post-infection, extracellular bacteria were removed by washing. At 24 h post-infection, intracellular bacteria were stabilized and collected in guanidine thiocyanate-based lysis buffer (4 M guanidine thiocyanate, 0.5% Na N-lauryl sarcosine, 25 mM sodium citrate and 0.1 M β-mercaptoethanol). Total RNA from was then extracted using a previously published method63.
RNA-Seq analysis
RNA samples were subjected to reverse transcription and the resulting cDNA was sequenced via Illumina HiSeq (100-bp paired end) at Macrogen. The resulting reads were first mapped to the H37Rv Mtb reference genome to retrieve bacterial sequences, resulting in 6–19 million bacterial sequence reads per sample. A strain-specific reference genome was prepared for each of the eight Mtb isolates, by polishing the H37Rv genome sequence with the Illumina reads previously obtained by sequencing whole genomic DNA extracted from that isolate (that is, the data used for phylogenomic analyses) using Pilon (v1.22) with default settings. For each isolate, the RNA-Seq reads were mapped to the isolate’s own reference genome sequence using Bowtie2 (v2.2.3)52. Read alignments were visualized using the BamView function in v14.0.064, which was also used to filter reads to include only those with a mapping score ≥30 for both forward and reverse reads, and to extract read counts for regions of interest. Expression values for the ESX-5 locus were calculated as the total reads mapping to the region spanning from pe19 to eccA5 (coordinates 2,029,880 to 2,038,532 in H37Rv). Due to the very close sequence homology in the protein-coding regions of the QILSS genes, it was not possible to assess their expression by counting total reads mapping to the length of the gene. Instead, we assessed the relative expression of these genes by extracting the number of reads mapping to a unique 30-bp marker region spanning the start codon of each paralog (coordinates −21 to +9 relative to the first base pair in each protein-coding sequence; see Fig. 4 for the uniqueness of these regions). To facilitate comparison of QILSS gene expression across strains with different RNA-Seq library sizes, we normalized the read counts for each isolate by the total number of reads mapping to the ESX-5 locus from pe19 to eccA5 (coordinates 2,029,880 to 2,038,532 in H37Rv). The ESX-5 locus was chosen for this purpose as all QILSS proteins are secreted by ESX-5 and thus depend on its expression (note that the locus contains the QILSS gene esxM, which was the highest expressed of all QILSS genes in all isolates). The ratios of reads mapping to each QILSS gene versus ESX-5 locus in each strain i (x i /ESX-5 i ) are shown in Fig. 4. To investigate differences in QILSS gene expression between wild-type (i) and mutant (j) isolate pairs (Fig. 4), we calculated the difference in these ratios relative to the wild-type level, as follows: (x i /ESX-5 i – x j /ESX-5 j )/(x i /ESX-5 i ).
Computational protein structure analysis
Structure-guided approaches can provide valuable insight into the molecular mechanism of mutations and their role in diseases20,65,66,67,68,69,70,71,72,73,74,75. To evaluate the structural effects of the Thr2Ala alteration in EsxW, models of EsxW and EsxV were generated using Modeller v9.1976 and MacroModel (vSchrodinger 2017, New York, NY), based on the experimental structures of homologous ESAT-6-like complexes (PDB IDs: 1WA8, 2KG7, 2VS0, 3OGI, 3ZBH, 4GZR, 4IOG and 4LWS; sequence identities ranging from 20–91%). The models were then minimized using the MMF94s forcefield in Sybyl-X 2.1.1 (Certara L.P., St Louis, MO), with the final structure having more than 95% of residues in the allowed region of a Ramachandran plot. The quality of the models was confirmed with Verify3D v1 (data not shown). Model structures were examined using Pymol v1.9. The model of the EsxW–EsxV complex was built using the homologous complexes to guide protein docking. The EsxW protomer and EsxW–EsxV heterodimer model were subjected to molecular dynamics to generate a series of representative conformations using Desmond (vSchrodinger 2017, New York, NY). Sodium and chloride ions were added to reach a final concentration of 150 mM Na+ and the system was solvated. The Amber and Charmm36 force fields were applied to the system and the default Desmond minimization and equilibration procedure was followed. Simulations were kept at constant pressure (1 atom) and temperature (335 K). The structural consequences of the EsxW-Thr2Ala variant were analyzed to account for all of the potential effects of the alterations77. The effects of the variant on the stability of EsxW were predicted using SDM v278, mCSM-Stability v179 and DUET v180. The effect of the alterations on the affinity of EsxW to bind to EsxV, and the stability of the complex, was predicted using mCSM-PPI v179. The predictions were analyzed and averaged across the conformational landscape. These computational approaches represent the wild-type structural and chemical environment of a residue as a graph-based signature to determine the change on alteration in Gibb’s free energy of stability or binding. Reported results are based on analysis of EsxW (wild-type and Thr2Ala mutant) and EsxV amino-acid sequences encoded in the H37Rv (lineage 4) reference genome; the only difference between these dimers and those encoded in lineage 1 or 2 is at EsxV residues 20 and 23. Structural modeling with the lineage 1/2 EsxV sequence yielded the same results as the analyses conducted with the lineage 4 background (that is, stronger binding affinity observed with the EsxW-Thr2Ala mutant).
Biophysical measurements
EsxV and wild-type (2Thr) and mutant (2Ala) EsxW were cloned into a pET18 vector with a carboxy-terminal hexahistidine tag (again using the sequences encoded in H37Rv (lineage 4) reference genome). A di-lysine motif was added before the hexahistidine tag of EsxW to facilitate labeling on the extended C terminus. Protein was expressed, purified and refolded following a previously published81. Solution microscale thermophoresis binding studies were performed to measure the binding affinity of EsxV for both the wild-type and mutant EsxW, using standard protocols on a Monolith NT.115 (Nanotemper Technologies)82,83,84. The EsxW was chosen to be labeled due to the presence of a lysine in EsxV at the heterodimer binding interface. As there were no lysines present in EsxW, the di-lysine motif was added to the C-terminal projection to facilitate labeling using a RED-NHS (Amine Reactive) Protein Labeling Kit (Nanotemper Technologies), which contains an NT-647 dye, as per the manufacturer’s instructions. Labeled wild-type or 2Ala mutant EsxW was mixed with EsxV in PBS with 0.05% Tween-20. Each replicate was undertaken using a 16-step twofold serial dilution series. The EsxW protein concentration was chosen such that the observed fluorescence was approximately 400 units at 70% light-emitting diode power. The samples were loaded into standard capillaries and heated at 40% laser power (48 mW) for 30 s, followed by 5 s cooling. The data were normalized against the baseline obtained in the absence of any EsxV, and the maximal response obtained at the highest concentration of inhibitor. The dissociation constant Kd was obtained by plotting the normalized fluorescence (Fnorm) against the logarithm of the concentrations of the dilution series and resulted in a sigmoidal binding curve that could be directly fitted with a nonlinear solution of the law of mass action (Fig. 4). All experiments were performed with four replicates, and the normalized fluorescence thermophoresis curves were analyzed using GraphPad v6 (GraphPad, San Diego, CA, USA).
Statistical analyses
All statistical analyses were performed in R version 3.3.3 unless stated otherwise. Associations between lineages and patient demographics (age group, sex) were assessed using Fisher’s exact test (two-sided in all cases) to assess n = 1,634 HCMC Mtb genomes (that is, excluding the lineage 3 isolate). A linear test for trend in proportions was also used to test for an increase in the frequency of lineage 1 within increasing age groups (also n = 1,634). Tests for difference in the distributions of terminal branch lengths or node-to-tip distances between lineages (Fig. 2) were conducted using the Kolmogorov–Smirnov test (two-sided in all cases, n = 1,634 Mtb genomes). Differences in the proportion of pairs involved in transmission clusters between lineages, or between case pairs isolated at the same DTU within ≤10 SNP or ≤20 SNP clusters versus pairs separated by >20 SNPs (Supplementary Fig. 1), were tested using a two-sided test for equal proportions (n = 1,634 Mtb genomes). A two-tailed t-test was used to compare Kd values for mutant versus wild-type (Fig. 4f), analysed using GraphPad Prism (n = 8; that is, 4 replicates each for wild-type and mutant).
URLs
RedDog pipeline v0.5, https://github.com/katholt/RedDog; European Nucleotide Archive, http://www.ebi.ac.uk/ena.
Reporting Summary
Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
Data availability
Mtb genome data were deposited in the NCBI BioProject database (ID: PRJNA355614); individual accession numbers for Mtb genomes analyzed in this study are given in Supplementary Tables 1 and 2 (including data from previous studies).
References
Zumla, A. et al. Eliminating tuberculosis and tuberculosis–HIV co-disease in the 21st century: key perspectives, controversies, unresolved issues, and needs. J. Infect. Dis. 205, S141–S146 (2012).
Global Tuberculosis Report 2017 (World Health Organization, 2017).
Casali, N. et al. Evolution and transmission of drug-resistant tuberculosis in a Russian population. Nat. Genet. 46, 279–286 (2014).
Guerra-Assuncao, J. A. et al. Large-scale whole genome sequencing of M. tuberculosis provides insights into transmission in a high prevalence area. eL ife https://doi.org/10.7554/eLife.05166 (2015).
Guerra-Assuncao, J. A. et al. Recurrence due to relapse or reinfection with Mycobacterium tuberculosis: a whole-genome sequencing approach in a large, population-based cohort with a high HIV infection prevalence and active follow-up. J. Infect. Dis. 211, 1154–1163 (2015).
Coll, F. et al. A robust SNP barcode for typing Mycobacterium tuberculosis complex strains. Nat. Commun. 5, 4812 (2014).
Anh, D. D. et al. Mycobacterium tuberculosis Beijing genotype emerging in Vietnam. Emerg. Infect. Dis. 6, 302–305 (2000).
Buu, T. N. et al. The Beijing genotype is associated with young age and multidrug-resistant tuberculosis in rural Vietnam. Int. J. Tuberc. Lung Dis. 13, 900–906 (2009).
Maeda, S. et al. Mycobacterium tuberculosis strains spreading in Hanoi, Vietnam: Beijing sublineages, genotypes, drug susceptibility patterns, and host factors. Tuberculosis 94, 649–656 (2014).
Nguyen, V. A. et al. High prevalence of Beijing and EAI4-VNM genotypes among M. tuberculosis isolates in northern Vietnam: sampling effect, rural and urban disparities. PLoS One 7, e45553 (2012).
Nguyen, V. A. et al. Mycobacterium tuberculosis lineages and anti-tuberculosis drug resistance in reference hospitals across Viet Nam. BMC Microbiol. 16, 167 (2016).
Horton, K. C., MacPherson, P., Houben, R. M., White, R. G. & Corbett, E. L. Sex differences in tuberculosis burden and notifications in low- and middle-income countries: a systematic review and meta-analysis. PLoS Med. 13, e1002119 (2016).
Malla, B. et al. First insights into the phylogenetic diversity of Mycobacterium tuberculosis in Nepal. PLoS One 7, e52297 (2012).
Lee, C. H. et al. Treatment delay and fatal outcomes of pulmonary tuberculosis in advanced age: a retrospective nationwide cohort study. BMC Infect. Dis. 17, 449 (2017).
Wlodarska, M., Johnston, J. C., Gardy, J. L. & Tang, P. A microbiological revolution meets an ancient disease: improving the management of tuberculosis with genomics. Clin. Microbiol. Rev. 28, 523–539 (2015).
Eldholm, V. et al. Four decades of transmission of a multidrug-resistant Mycobacterium tuberculosis outbreak strain. Nat. Commun. 6, 7119 (2015).
Zhang, H. et al. Genome sequencing of 161 Mycobacterium tuberculosis isolates from China identifies genes and intergenic regions associated with drug resistance. Nat. Genet. 45, 1255–1260 (2013).
Merker, M. et al. Evolutionary history and global spread of the Mycobacterium tuberculosis Beijing lineage. Nat. Genet. 47, 242–249 (2015).
Comas, I. et al. Population genomics of Mycobacterium tuberculosis in Ethiopia contradicts the virgin soil hypothesis for human tuberculosis in sub-Saharan Africa. Curr. Biol. 25, 3260–3266 (2015).
Phelan, J. et al. Mycobacterium tuberculosis whole genome sequencing and protein structure modelling provides insights into anti-tuberculosis drug resistance. BMC Med. 14, 31 (2016).
Stucki, D. et al. Mycobacterium tuberculosis lineage 4 comprises globally distributed and geographically restricted sublineages. Nat. Genet. 48, 1535–1543 (2016).
Hanekom, M. et al. Mycobacterium tuberculosis Beijing genotype: a template for success. Tuberculosis 91, 510–523 (2011).
Parwati, I., van Crevel, R. & van Soolingen, D. Possible underlying mechanisms for successful emergence of the Mycobacterium tuberculosis Beijing genotype strains. Lancet Infect. Dis. 10, 103–111 (2010).
Coscolla, M. & Gagneux, S. Consequences of genomic diversity in Mycobacterium tuberculosis. Semin. Immunol. 26, 431–44 (2014).
van Laarhoven, A. et al. Low induction of proinflammatory cytokines parallels evolutionary success of modern strains within the Mycobacterium tuberculosis Beijing genotype. Infect. Immun. 81, 3750–3756 (2013).
Farhat, M. R. et al. Genomic analysis identifies targets of convergent positive selection in drug-resistant Mycobacterium tuberculosis. Nat. Genet. 45, 1183–1189 (2013).
Hazbon, M. H. et al. Convergent evolutionary analysis identifies significant mutations in drug resistance targets of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 52, 3369–3376 (2008).
Knudsen, N. P. et al. Tuberculosis vaccine with high predicted population coverage and compatibility with modern diagnostics. Proc. Natl Acad. Sci. USA 111, 1096–1101 (2014).
Baldwin, S. L. et al. Intradermal immunization improves protective efficacy of a novel TB vaccine candidate. Vaccine 27, 3063–3071 (2009).
Baldwin, S. L. et al. Protection and long-lived immunity induced by the ID93/GLA-SE vaccine candidate against a clinical Mycobacterium tuberculosis isolate. Clin. Vaccine Immunol. 23, 137–147 (2015).
Bertholet, S. et al. A defined tuberculosis vaccine candidate boosts BCG and protects against multidrug-resistant Mycobacterium tuberculosis. Sci. Transl. Med. 2, 53ra74 (2010).
Bertholet, S. et al. Identification of human T cell antigens for the development of vaccines against Mycobacterium tuberculosis. J. Immunol. 181, 7948–7957 (2008).
Uplekar, S., Heym, B., Friocourt, V., Rougemont, J. & Cole, S. T. Comparative genomics of Esx genes from clinical isolates of Mycobacterium tuberculosis provides evidence for gene conversion and epitope variation. Infect. Immun. 79, 4042–4049 (2011).
Lewinsohn, D. M. et al. Human Mycobacterium tuberculosis CD8 T cell antigens/epitopes identified by a proteomic peptide library. PLoS One 8, e67016 (2013).
Kim, Y. et al. Immune epitope database analysis resource. Nucleic Acids Res. 40, W525–W530 (2012).
Mortimer, T. D., Weber, A. M. & Pepperell, C. S. Evolutionary thrift: Mycobacteria repurpose plasmid diversity during adaptation of type VII secretion systems. Genome Biol. Evol. 9, 398–413 (2017).
Gey Van Pittius, N. C. et al. The ESAT-6 gene cluster of Mycobacterium tuberculosis and other high G + C Gram-positive bacteria. Genome Biol. 2, RESEARCH0044 (2001).
Abdallah, A. M. et al. PPE and PE_PGRS proteins of Mycobacterium marinum are transported via the type VII secretion system ESX-5. Mol. Microbiol. 73, 329–340 (2009).
Fishbein, S., van Wyk, N., Warren, R. M. & Sampson, S. L. Phylogeny to function: PE/PPE protein evolution and impact on Mycobacterium tuberculosis pathogenicity. Mol. Microbiol. 96, 901–916 (2015).
Brennan, M. J. The enigmatic PE/PPE multigene family of Mycobacteria and tuberculosis vaccination. Infect. Immun. 85, e00969-16 (2017).
Groschel, M. I., Sayes, F., Simeone, R., Majlessi, L. & Brosch, R. ESX secretion systems: mycobacterial evolution to counter host immunity. Nat. Rev. Microbiol. 14, 677–691 (2016).
Shah, S., Cannon, J. R., Fenselau, C. & Briken, V. A duplicated ESAT-6 region of ESX-5 is involved in protein export and virulence of Mycobacteria. Infect. Immun. 83, 4349–4361 (2015).
Kumar, A., Chandolia, A., Chaudhry, U., Brahmachari, V. & Bose, M. Comparison of mammalian cell entry operons of Mycobacteria: in silico analysis and expression profiling. FEMS Immunol. Med. Microbiol. 43, 185–195 (2005).
Bukka, A., Price, C. T., Kernodle, D. S. & Graham, J. E. Mycobacterium tuberculosis RNA expression patterns in sputum bacteria indicate secreted Esx factors contributing to growth are highly expressed in active disease. Front. Microbiol. 2, 266 (2012).
Coppola, M. et al. New genome-wide algorithm identifies novel in-vivo expressed mycobacterium tuberculosis antigens inducing human T-cell responses with classical and unconventional cytokine profiles. Sci. Rep. 6, 37793 (2016).
Abdallah, A. M. et al. Mycobacterial secretion systems ESX-1 and ESX-5 play distinct roles in host cell death and inflammasome activation. J. Immunol. 187, 4744–4753 (2011).
Ford, C. B. et al. Mycobacterium tuberculosis mutation rate estimates from different lineages predict substantial differences in the emergence of drug-resistant tuberculosis. Nat. Genet. 45, 784–790 (2013).
Fox, G. J., Barry, S. E., Britton, W. J. & Marks, G. B. Contact investigation for tuberculosis: a systematic review and meta-analysis. Eur. Respir. J. 41, 140–156 (2013).
Lonnroth, K. et al. Systematic screening for active tuberculosis: rationale, definitions and key considerations. Int. J. Tuberc. Lung Dis. 17, 289–298 (2013).
Thai, P. V. K. et al. Bacterial risk factors for treatment failure and relapse among patients with isoniazid resistant tuberculosis. BMC Infect. Dis. 18, 112 (2018).
Caws, M. et al. The influence of host and bacterial genotype on the development of disseminated disease with Mycobacterium tuberculosis. PLoS Pathog. 4, e1000034 (2008).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–9 (2012).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Comas, I. et al. Out-of-Africa migration and Neolithic coexpansion of Mycobacterium tuberculosis with modern humans. Nat. Genet. 45, 1176–1182 (2013).
Pepperell, C. S. et al. The role of selection in shaping diversity of natural M. tuberculosis populations. PLoS Pathog. 9, e1003543 (2013).
Bradley, P. et al. Rapid antibiotic-resistance predictions from genome sequence data for Staphylococcus aureus and Mycobacterium tuberculosis. Nat. Commun. 6, 10063 (2015).
Stamatakis, A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014).
Price, M. N., Dehal, P. S. & Arkin, A. P. FastTree 2 – approximately maximum-likelihood trees for large alignments. PLoS One 5, e9490 (2010).
Ashkenazy, H. et al. FastML: a web server for probabilistic reconstruction of ancestral sequences. Nucleic Acids Res. 40, W580–W584 (2012).
Bollback, J. P. SIMMAP: stochastic character mapping of discrete traits on phylogenies. BMC Bioinformatics 7, 88 (2006).
Revell, L. phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 3, 217–223 (2012).
Vijay, S. V. et al. Influence of stress and antibiotic resistance on cell-length distribution in Mycobacterium tuberculosis clinical isolates. Front. Microbiol. 8, 1–12 (2017).
Rohde, K. H., Abramovitch, R. B. & Russell, D. G. Mycobacterium tuberculosis invasion of macrophages: linking bacterial gene expression to environmental cues. Cell Host Microbe 2, 352–364 (2007).
Carver, T. et al. BamView: visualizing and interpretation of next-generation sequencing read alignments. Brief Bioinform. 14, 203–212 (2013).
Jafri, M. et al. Germline mutations in the CDKN2B tumor suppressor gene predispose to renal cell carcinoma. Cancer Discov. 5, 723–729 (2015).
Usher, J. L. et al. Analysis of HGD gene mutations in patients with alkaptonuria from the United Kingdom: identification of novel mutations. JIMD Rep. 24, 3–11 (2015).
Jubb, H. C. et al. Mutations at protein–protein interfaces: Small changes over big surfaces have large impacts on human health. Prog. Biophys. Mol. Biol. 128, 3–13 (2017).
Kano, F. S. et al. The presence, persistence and functional properties of Plasmodium vivax Duffy binding protein II antibodies are influenced by HLA class II allelic variants. PLoS Negl. Trop. Dis. 10, e0005177 (2016).
Nemethova, M. et al. Twelve novel HGD gene variants identified in 99 alkaptonuria patients: focus on ‘black bone disease’ in Italy. Eur. J. Hum. Genet. 24, 66–72 (2016).
Silvino, A. C. et al. Variation in human cytochrome P-450 drug-metabolism genes: a gateway to the understanding of Plasmodium vivax relapses. PLoS One 11, e0160172 (2016).
White, R. R. et al. Ubiquitin-dependent modification of skeletal muscle by the parasitic nematode, Trichinella spiralis. PLoS Pathog. 12, e1005977 (2016).
Albanaz, A. T. S., Rodrigues, C. H. M., Pires, D. E. V. & Ascher, D. B. Combating mutations in genetic disease and drug resistance: understanding molecular mechanisms to guide drug design. Expert Opin. Drug Discov. 12, 553–563 (2017).
Casey, R. T. et al. SDHA related tumorigenesis: a new case series and literature review for variant interpretation and pathogenicity. Mol. Genet. Genomic Med. 5, 237–250 (2017).
Pandurangan, A. P., Ascher, D. B., Thomas, S. E. & Blundell, T. L. Genomes, structural biology and drug discovery: combating the impacts of mutations in genetic disease and antibiotic resistance. Biochem. Soc. Trans. 45, 303–311 (2017).
Soardi, F. C. et al. Familial STAG2 germline mutation defines a new human cohesinopathy. NPJ Genom. Med. https://doi.org/10.1038/s41525-017-0009-4 (2017).
Sali, A. & Blundell, T. L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815 (1993).
Pires, D. E., Chen, J., Blundell, T. L. & Ascher, D. B. In silico functional dissection of saturation mutagenesis: interpreting the relationship between phenotypes and changes in protein stability, interactions and activity. Sci. Rep. 6, 19848 (2016).
Pandurangan, A. P., Ochoa-Montano, B., Ascher, D. B. & Blundell, T. L. SDM: a server for predicting effects of mutations on protein stability. Nucleic Acids Res. 45(W1), W229–W235 (2017).
Pires, D. E., Ascher, D. B. & Blundell, T. L. mCSM: predicting the effects of mutations in proteins using graph-based signatures. Bioinformatics 30, 335–42 (2014).
Pires, D. E., Ascher, D. B. & Blundell, T. L. DUET: a server for predicting effects of mutations on protein stability using an integrated computational approach. Nucleic Acids Res. 42, W314–W319 (2014).
Mahmood, A. et al. Molecular characterization of secretory proteins Rv3619c and Rv3620c from Mycobacterium tuberculosis H37Rv. FEBS J. 278, 341–53 (2011).
Ascher, D. B. et al. Potent hepatitis C inhibitors bind directly to NS5A and reduce its affinity for RNA. Sci. Rep. 4, 4765 (2014).
Chan, L. J. et al. Conjugation of 10 kDa linear PEG onto trastuzumab Fab’ is sufficient to significantly enhance lymphatic exposure while preserving in vitro biological activity. Mol. Pharm. 13, 1229–1241 (2016).
Chan, L. J. et al. PEGylation does not significantly change the initial intravenous or subcutaneous pharmacokinetics or lymphatic exposure of trastuzumab in rats but increases plasma clearance after subcutaneous administration. Mol. Pharm. 12, 794–809 (2015).
Acknowledgements
We would like to thank the clinical staff who recruited patients into our study from the following District TB Units (DTUs) in HCMC, Vietnam: Districts 1, 4, 5, 6 and 8, Tan Binh, Binh Thanh and Phu Nhuan; and also our colleagues from Pham Ngoc Thach Hospital for Tuberculosis and Lung Disease, HCMC Vietnam. This work was supported by the National Health and Medical Research Council, Australia (project grant no. 1056689 to S.J.D., Fellowship no. 1061409 to K.E.H., Fellowship no. 1061435 to M.I., Fellowship no. 1072476 to D.B.A.), A*STAR Biomedical Research Council, Singapore (12/1/21/24/6689 to Y.Y.T.) and the Wellcome Trust UK (research training fellowship no. 081814/Z/06/Z to M.C.) and as part of their Major Overseas Program in Vietnam (089276/Z/09/Z to J.F. and 106680/B/14/Z to G.T.).
Author information
Authors and Affiliations
Contributions
S.J.D., K.E.H., M.C., M.I., Y.Y.T. and C.C.K. are the study principal investigators who conceived and obtained funding for the project. S.J.D. provided overall project coordination; M.I. organized and supervised the DNA sequencing and K.E.H. devised the overall analysis plan and wrote the first draft of the manuscript along with P.M. M.C. and S.J.D. established the TB cohort for this genetics study by working with P.V.K.T., D.T.M.H., N.N.L., N.H.L., N.T.Q.N., N.T.T.T., G.T. and J.F. to coordinate the collection of clinical samples and phenotypes. K.P. performed DNA quality checks and genome sequencing on all Vietnamese samples, while V.T.N.H. performed Sanger sequencing on selected samples. D.B.A. performed protein structure analyses, and H.T.H. and N.T.T.T. performed the macrophage growth and infection experiments of EsxW variants. K.E.H., P.M., M.I., D.J.E. and A.P.N. analyzed the data. All authors critically reviewed manuscript revisions and contributed intellectual input to the final submission.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 Association between Mtb genetic distance and spatial proximity of patients.
For all pairs of Mtb isolates, the probability of both originating from patients living in the same district was calculated (y-axis) and stratified according to genetic distance between the Mtb genomes (expressed in number of SNPs, x-axis). P-values indicate result of testing for difference in proportions of isolate pairs originating from same district, for isolate pairs with SNP distance <10 vs >20, calculated separately for Beijing and non-Beijing isolates. (Note it was not possible to further stratify non-Beijing pairs into L1 pairs vs L4 pairs for statistical tests, due to low sample size.).
Supplementary Figure 2 Proportion of clusters not explained by transmission of antimicrobial resistance mutations.
Each cluster (defined at various thresholds of maximum patristic distance between tips) was checked to determine whether all members of the cluster shared any of the antimicrobial resistance mutations identified by Mykrobe Predictor. Clusters in which no known antimicrobial resistance mutation was conserved in all members of the cluster were considered not driven by antimicrobial resistance. For each lineage and at each maximum patristic distance threshold value for clustering, the proportion of clusters not explained by antimicrobial resistance are shown below. Colour legend for lineages is the same as throughout the paper: pink, lineage 1; dark blue, lineage 2.1; orange, lineage 2.2.1; light blue, lineage 2.2.2; red, lineage 4.
Supplementary Figure 3 Homoplasic mutations detected in the HCMC Mtb population and located in known drug resistance-associated genes.
Mutations arising on more than one branch in the HCMC Mtb tree in the following genes are included: embB (ethambutol resistance), gidB (streptomycin resistance), gyrA (fluoroquinolone resistance); inhA (isoniazid resistance), rpoB (rifampicin resistance).
Supplementary Figure 4 Homoplasic nsSNPs that are fixed within Beijing lineage and also arise independently in other branches in the HCMC phylogeny.
Tree shown is the ML phylogeny for 1,635 HCMC isolates with lineages highlighted, reproduced from Figure 1. Outer rings indicate the presence of derived alleles for the three nsSNPs that were found to be fixed within the Beijing lineage (2.2; including 2.2.1 and 2.2.2).
Supplementary Figure 5 Map of HCMC showing location of district TB units where patients were recruited.
Yellow regions show districts included in the study; red dots indicate location of the district TB units where participants were recruited; red star indicates location of Pham Ngoc Thach Hospital for Tuberculosis and Lung Disease. Blue regions indicate other urban HCMC districts, green are greater HCMC districts (not included in recruitment).
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–5 and Supplementary Tables 3 and 4
Supplementary Table 1
Details of the M. tuberculosis isolates sequenced for this study
Supplementary Table 2
Details of the publicly available M. tuberculosis genomes included in this study
Supplementary Table 5
Repetitive regions of the M. tuberculosis genome that were excluded from SNP analysis
Rights and permissions
About this article
Cite this article
Holt, K.E., McAdam, P., Thai, P.V.K. et al. Frequent transmission of the Mycobacterium tuberculosis Beijing lineage and positive selection for the EsxW Beijing variant in Vietnam. Nat Genet 50, 849–856 (2018). https://doi.org/10.1038/s41588-018-0117-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-018-0117-9
This article is cited by
-
Parallel signatures of Mycobacterium tuberculosis and human Y-chromosome phylogeography support the Two Layer model of East Asian population history
Communications Biology (2023)
-
Evolution and spread of a highly drug resistant strain of Mycobacterium tuberculosis in Papua New Guinea
BMC Infectious Diseases (2022)
-
Whole-genome single nucleotide variant phylogenetic analysis of Mycobacterium tuberculosis Lineage 1 in endemic regions of Asia and Africa
Scientific Reports (2022)
-
A genome epidemiological study of mycobacterium tuberculosis in subpopulations with high and low incidence rate in Guangxi, South China
BMC Infectious Diseases (2021)
-
Assessment of closely related Mycobacterium tuberculosis variants with different transmission success and in vitro infection dynamics
Scientific Reports (2021)