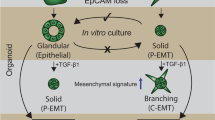
Abstract
Pancreatic ductal adenocarcinoma (PDAC) shows pronounced epithelial and mesenchymal cancer cell populations1,2,3,4. Cellular heterogeneity in PDAC is an important feature in disease subtype specification3,4,5, but how distinct PDAC subpopulations interact, and the molecular mechanisms that underlie PDAC cell fate decisions, are incompletely understood. Here we identify the BMP inhibitor GREM16,7 as a key regulator of cellular heterogeneity in pancreatic cancer in human and mouse. Grem1 inactivation in established PDAC in mice resulted in a direct conversion of epithelial into mesenchymal PDAC cells within days, suggesting that persistent GREM1 activity is required to maintain the epithelial PDAC subpopulations. By contrast, Grem1 overexpression caused an almost complete ‘epithelialization’ of highly mesenchymal PDAC, indicating that high GREM1 activity is sufficient to revert the mesenchymal fate of PDAC cells. Mechanistically, Grem1 was highly expressed in mesenchymal PDAC cells and inhibited the expression of the epithelial–mesenchymal transition transcription factors Snai1 (also known as Snail) and Snai2 (also known as Slug) in the epithelial cell compartment, therefore restricting epithelial–mesenchymal plasticity. Thus, constant suppression of BMP activity is essential to maintain epithelial PDAC cells, indicating that the maintenance of the cellular heterogeneity of pancreatic cancer requires continuous paracrine signalling elicited by a single soluble factor.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
RNA-seq data for purified EPCAM+ and EPCAM− KPCY tumour cells and for KPFCT;Grem1WT/WT and KPFCT;Grem1∆/∆ tumour tissues have been deposited in the Gene Expression Omnibus under the accession codes GSE157134 and GSE182143, respectively. The scRNA-seq datasets for the 24 PDAC and 11 normal pancreas samples were downloaded from the Chinese National Genomics Data Center (Genome Sequence Archive: CRA001160)53. RNA-seq data for human pancreatic cancer cell lines were downloaded from DepMap portal (expression 21Q2 public; https://doi.org/10.6084/m9.figshare.14541774.v2)60. Microarray data for GREM1 in human PDACs were downloaded from NCBI GEO dataset (GSE71729)18. Clinical data and normalized RSEM read counts for the PDAC TCGA dataset were downloaded from the Broad firehose data repository (version 2016_01_28; https://gdac.broadinstitute.org/runs/stddata__2016_01_28/data/PAAD/20160128/). The mouse reference genome GRCm38 was obtained from Ensembl version 8642 or 10245 (http://nov2020.archive.ensembl.org/Mus_musculus/Info/Index). Source data are provided with this paper.
References
Wang, V. M. et al. CD9 identifies pancreatic cancer stem cells and modulates glutamine metabolism to fuel tumour growth. Nat. Cell Biol. 21, 1425–1435 (2019).
Ligorio, M. et al. Stromal microenvironment shapes the intratumoral architecture of pancreatic cancer. Cell 178, 160–175 (2019).
Chan-Seng-Yue, M. et al. Transcription phenotypes of pancreatic cancer are driven by genomic events during tumor evolution. Nat. Genet. 52, 231–240 (2020).
Hayashi, A. et al. A unifying paradigm for transcriptional heterogeneity and squamous features in pancreatic ductal adenocarcinoma. Nat. Cancer 1, 59–74 (2020).
Collisson, E. A., Bailey, P., Chang, D. K. & Biankin, A. V. Molecular subtypes of pancreatic cancer. Nat. Rev. Gastroenterol. Hepatol. 16, 207–220 (2019).
Brazil, D. P., Church, R. H., Surae, S., Godson, C. & Martin, F. BMP signalling: agony and antagony in the family. Trends Cell Biol. 25, 249–264 (2015).
Ouahoud, S., Hardwick, J. C. H. & Hawinkels, L. Extracellular BMP antagonists, multifaceted orchestrators in the tumor and its microenvironment. Int. J. Mol. Sci. 21, 3888 (2020).
Sankpal, N. V., Fleming, T. P., Sharma, P. K., Wiedner, H. J. & Gillanders, W. E. A double-negative feedback loop between EPCAM and ERK contributes to the regulation of epithelial-mesenchymal transition in cancer. Oncogene 36, 3706–3717 (2017).
Pastushenko, I. et al. Identification of the tumour transition states occurring during EMT. Nature 556, 463–468 (2018).
Bardeesy, N. et al. Both p16Ink4a and the p19Arf-p53 pathway constrain progression of pancreatic adenocarcinoma in the mouse. Proc. Natl Acad. Sci. USA 103, 5947–5952 (2006).
Schönhuber, N. et al. A next-generation dual-recombinase system for time- and host-specific targeting of pancreatic cancer. Nat. Med. 20, 1340–1347 (2014).
Gazzerro, E. et al. Conditional deletion of gremlin causes a transient increase in bone formation and bone mass. J. Biol. Chem. 282, 31549–31557 (2007).
Muzumdar, M. D., Tasic, B., Miyamichi, K., Li, L. & Luo, L. A global double-fluorescent Cre reporter mouse. Genesis 45, 593–605 (2007).
Collisson, E. A. et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat. Med. 17, 500–503 (2011).
Chaffer, C. L., San Juan, B. P., Lim, E. & Weinberg, R. A. EMT, cell plasticity and metastasis. Cancer Metastasis Rev. 35, 645–654 (2016).
Dongre, A. & Weinberg, R. A. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat. Rev. Mol. Cell Biol. 20, 69–84 (2019).
Yang, J. et al. Guidelines and definitions for research on epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 21, 341–352 (2020).
Moffitt, R. A. et al. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat. Genet. 47, 1168–1178 (2015).
Neuzillet, C. et al. Inter- and intra-tumoural heterogeneity in cancer-associated fibroblasts of human pancreatic ductal adenocarcinoma. J. Pathol. 248, 51–65 (2019).
Pastushenko, I. & Blanpain, C. EMT transition states during tumor progression and metastasis. Trends Cell Biol. 29, 212–226 (2019).
Sanvitale, C. E. et al. A new class of small molecule inhibitor of BMP signaling. PLoS ONE 8, e62721 (2013).
Stemmler, M. P., Eccles, R. L., Brabletz, S. & Brabletz, T. Non-redundant functions of EMT transcription factors. Nat. Cell Biol. 21, 102–112 (2019).
Guaita, S. et al. Snail induction of epithelial to mesenchymal transition in tumor cells is accompanied by MUC1 repression and ZEB1 expression. J. Biol. Chem. 277, 39209–39216 (2002).
Tran, D. D., Corsa, C. A., Biswas, H., Aft, R. L. & Longmore, G. D. Temporal and spatial cooperation of Snail1 and Twist1 during epithelial-mesenchymal transition predicts for human breast cancer recurrence. Mol. Cancer Res. 9, 1644–1657 (2011).
Dave, N. et al. Functional cooperation between Snail1 and twist in the regulation of ZEB1 expression during epithelial to mesenchymal transition. J. Biol. Chem. 286, 12024–12032 (2011).
Kröger, C. et al. Acquisition of a hybrid E/M state is essential for tumorigenicity of basal breast cancer cells. Proc. Natl Acad. Sci. USA 116, 7353–7362 (2019).
Turing, A. M. The chemical basis of morphogenesis. Philos. Trans. R. Soc. Lond. B 237, 37–72 (1952).
Madamanchi, A., Mullins, M. C. & Umulis, D. M. Diversity and robustness of bone morphogenetic protein pattern formation. Development 148, dev192344 (2021).
Jackson, E. L. et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 15, 3243–3248 (2001).
Marino, S., Vooijs, M., van Der Gulden, H., Jonkers, J. & Berns, A. Induction of medulloblastomas in p53-null mutant mice by somatic inactivation of Rb in the external granular layer cells of the cerebellum. Genes Dev. 14, 994–1004 (2000).
Hingorani, S. R. et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell 4, 437–450 (2003).
Srinivas, S. et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 1, 4 (2001).
Flanagan, S. P. ‘Nude’, a new hairless gene with pleiotropic effects in the mouse. Genet. Res. 8, 295–309 (1966).
Wang, F. et al. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J. Mol. Diagn. 14, 22–29 (2012).
Boj, S. F. et al. Organoid models of human and mouse ductal pancreatic cancer. Cell 160, 324–338 (2015).
Yusa, K., Zhou, L., Li, M. A., Bradley, A. & Craig, N. L. A hyperactive piggyBac transposase for mammalian applications. Proc. Natl Acad. Sci. USA 108, 1531–1536 (2011).
Fujii, M., Matano, M., Nanki, K. & Sato, T. Efficient genetic engineering of human intestinal organoids using electroporation. Nat. Protoc. 10, 1474–1485 (2015).
Ramachandran, A. et al. TGF-β uses a novel mode of receptor activation to phosphorylate SMAD1/5 and induce epithelial-to-mesenchymal transition. eLife 7, e31756 (2018).
Kechin, A., Boyarskikh, U., Kel, A. & Filipenko, M. cutPrimers: a new tool for accurate cutting of primers from reads of targeted next generation sequencing. J. Comput. Biol. 24, 1138–1143 (2017).
Li, B. & Dewey, C. N. RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinform. 12, 323 (2011).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Aken, B. L. et al. Ensembl 2017. Nucleic Acids Res. 45, D635–D642 (2017).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Kim, D., Paggi, J. M., Park, C., Bennett, C. & Salzberg, S. L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915 (2019).
Howe, K. L. et al. Ensembl 2021. Nucleic Acids Res. 49, D884–D891 (2021).
Jolliffe, I. T. & Cadima, J. Principal component analysis: a review and recent developments. Philos. Trans. A 374, 20150202 (2016).
Durinck, S. et al. BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics 21, 3439–3440 (2005).
Hoshida, Y. Nearest template prediction: a single-sample-based flexible class prediction with confidence assessment. PLoS ONE 5, e15543 (2010).
Reich, M. et al. GenePattern 2.0. Nat. Genet. 38, 500–501 (2006).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Elyada, E. et al. Cross-species single-cell analysis of pancreatic ductal adenocarcinoma reveals antigen-presenting cancer-associated fibroblasts. Cancer Discov. 9, 1102–1123 (2019).
Liberzon, A. et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425 (2015).
Kanehisa, M. & Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 28, 27–30 (2000).
Ashburner, M. et al. Gene Ontology: tool for the unification of biology. Nat. Genet. 25, 25–29 (2000).
Peng, J. et al. Single-cell RNA-seq highlights intra-tumoral heterogeneity and malignant progression in pancreatic ductal adenocarcinoma. Cell Res. 29, 725–738 (2019).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018).
Liberzon, A. et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics 27, 1739–1740 (2011).
Huang da, W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009).
Fornes, O. et al. JASPAR 2020: update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 48, D87–d92 (2020).
Ghandi, M. et al. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature 569, 503–508 (2019).
Acknowledgements
We thank D. Saur for providing the Pdx1-Flp, KrasFSF-G12D, Trp53frt and Rosa26FSF-creER mouse lines; P. Srivastava and J. Ji for assisting scRNA-seq data acquisition and processing; G. Salbreux for discussions about the Turing mechanism; and the staff at the Francis Crick Institute Biological Research, Experimental Histopathology, Flow Cytometry, Advanced Sequencing and Light Microscopy facilities for technical support. This work was supported by the Francis Crick Institute, which receives its core funding from Cancer Research UK (FC001039, FC001317), the UK Medical Research Council (FC001039, FC001317) and the Wellcome Trust (FC001039, FC001317), and by the Breast Cancer Now Toby Robins Research Centre at the Institute of Cancer Research (CTR-Q5-Y2). L.L. was supported by a Mildred-Scheel-Postdoctoral stipend (70112263) from German Cancer Aid (04/2016-03/2017) and by a research fellowship (LA 3927/1) from German Research Foundation (04/2017–03/2020).
Author information
Authors and Affiliations
Contributions
L.L. and A.B. conceived and designed the study and wrote the manuscript. L.L. performed the experiments and analyses. T.E. helped with RNA-seq, stromal cell analysis and organoid electroporation experiments. H.L. helped with organoid cultures, organoid IF and qPCR analysis. A.H., H.P. and A.S. performed human PDAC subtype correlation and human PDAC scRNA-seq analysis. M.Z.T. helped with ultrasound imaging of pancreatic tumours. R.M.M.F. and E.M.R. assisted with study design. Y.Z. assisted with histopathological evaluation. J.A. helped with lentiviral transduction and subcutaneous injections. K.N. performed flow cytometry analysis of immune infiltration. P.S.-B. assisted with the RNA-seq analysis. J.N. managed the KPCY colony and assisted with KPCY tumour tissue sections preparation. E.J.R. and G.K. assisted with data interpretation and analysis and text proofing. J.C. and E.L.N. performed the RNAscope and immunohistochemistry experiments. Y.H. and C.Z. provided intellectual feedback. A.B. supervised the study.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Grem1 is upregulated in EPCAM− tumour cells in pancreatic cancer.
a, b, Representative images for immunofluorescent staining of EPCAM, YFP, CK19 (a) and vimentin (b) on KPCY tumours (n = 3 mice). Scale bar, 100 μm. Arrowheads indicate CK19+ tumour cells that lost EPCAM expression. c, Heatmap for the hallmark EMT gene set that is enriched in EPCAM− tumour cells. Some mesenchymal associated genes, including Grem1 are displayed. d, The table for functional annotations of significantly upregulated genes in EPCAM− tumour cells by the DAVID database. Top 2 enriched functional clusters are shown. ES, enrichment score. The P values (EASE scores) are from Fisher's exact test. e, f, Representative images for RNAscope staining of Grem1 and IF staining of YFP and vimentin in early KPCY tumour lesions (n = 3 mice). Scale bar, 100 μm. Indicated areas are magnified on the right. Scale bar, 25 μm.
Extended Data Fig. 2 GREM1 expression highly correlates with EMT in human pancreatic cancer.
a, Normalized RNA read counts (RSEM) for GREM1 from the TCGA PDAC dataset. The samples were categorized into GREM1hi and GREM1lo groups based on its expression, higher or lower than the median value. The lines in violin plots show the median, the upper quartile and the lower quartile respectively. P value is from two-sided Mann-Whitney test. b, GSEA plot for enrichment of the hallmark EMT gene set in the transcriptome of GREM1hi PDACs. NES, normalized enrichment score; FDR, false discovery rate. c, RSEM for epithelial and mesenchymal genes and GREM1 in the Cancer Cell Line Encyclopedia dataset for human PDAC cell lines. Cell lines were categorized into VIMhi and VIMlo groups based on its expression higher or lower than the median value. The lines in violin plots show the median, the upper quartile and the lower quartile respectively. P values are from two-sided t-test. d, UMAP plot of single cells in a scRNA-seq dataset for human PDACs (n = 24 biologically independent samples). Ductal cell type 1 and 2 represent two main cancer cell populations. e, Cancer cells were categorized into GREM1pos (n = 410 cells) and GREM1neg (n = 13551 cells) groups based on the expression of GREM1. The box and the line in the center show the interquartile range (between the 25th and 75th percentile) and the median respectively, and whiskers extend to the largest and smallest values within 1.5 times the interquartile range. P value is from two-sided Wilcoxon test. f, EMT signature score plot for GREM1pos and GREM1neg cancer cells. P value is from two-sided Wilcoxon test.
Extended Data Fig. 3 KPCY and KPFCT pancreatic tumours show similar EMT features and Grem1 expression pattern.
a, Immunofluorescent staining of YFP, CK19, and vimentin in KPCY tumours. Scale bar, 200 μm. Indicated areas are magnified on the right. Scale bar, 50 μm. b, Quantification of the number of CK19+vimentin−, CK19+vimentin− and CK19+vimentin− KPCY tumour cells (n = 5 mice). c, Immunofluorescent staining of GFP, CK19 and vimentin in KPFCT tumours. Scale bar, 200 μm. Indicated areas are magnified on the right. Scale bar, 50 μm. d, Quantification of the number of CK19+vimentin−, CK19+vimentin− and CK19+vimentin− KPFCT tumour cells (n = 5 mice). e, RNAscope staining of Grem1 and IF staining of GFP and vimentin in KPFCT tumours. Scale bar, 100 μm. Indicated areas are magnified on the right and bottom. Scale bar, 30 μm. Arrowheads indicates Grem1-expressing tumour cells. f, Quantification of the percentage of epithelial tumour cells (ETCs), mesenchymal tumour cells (MTCs) and stromal cells (SCs) in the Grem1+ population (n = 3 mice). The bar charts show the mean ± s.e.m. P values are from one-way ANOVA and Tukey multiple comparison test.
Extended Data Fig. 4 Grem1 deletion induces highly mesenchymal pancreatic tumours.
a, Ultrasound imaging of pancreatic tumours in 5-week-old KPFCT mice. The dashed red circles indicate the tumours in the pancreas and the numbers denote the tumour volume. b, Quantification of the proportion of 5-week-old KPFCT mice with tumours at different sizes (n = 8). c, H&E staining of a small and large tumour from 5-week-old KPFCT mice (n = 8). Scale bar, 200 μm. d, Kaplan–Meier survival plot for KPFCT/Grem1wt/wt (n = 10), KPFCT/Grem1wt/Δ (n = 10) and KPFCT/Grem1Δ/Δ (n = 7) mice that received tamoxifen injections at 5 weeks of age. e, RNAscope staining of Grem1 and IHC staining of GFP in KPFCT/Grem1wt/wt and KPFCT/Grem1Δ/Δ tumours. Scale bar, 50 μm. f, RT–qPCR analysis of Grem1 on FACS-sorted GFP+ cells from KPFCT/Grem1wt/wt and KPFCT/Grem1Δ/Δ tumours (n = 3 mice). The expression was normalized to Rplp0, and the relative level between samples was calculated using the 2^(-∆∆Ct) method. The bar charts show the mean ± s.e.m. g, H&E and IHC staining for CK19 in KPFCT/Grem1wt/wt and KPFCT/Grem1Δ/Δ tumours, showing Grem1 deletion induced highly mesenchymal pancreatic tumours (n = 5 mice). Scale bar, 2000 μm. Indicated areas are magnified on the right. Scale bar, 100 μm.
Extended Data Fig. 5 Grem1-deficient tumours organoids show EMT features and Grem1-deficient tumours correlate with the quasi-mesenchymal subtype of human PDAC.
a, Scheme depicting the experimental approach for establishing KPFCT/Grem1wt/wt and KPFCT/Grem1Δ/Δ organoids from tamoxifen-treated tumours. b, RT–qPCR analysis of epithelial and mesenchymal genes on KPFCT/Grem1wt/wt and KPFCT/Grem1Δ/Δ organoids (n = 3 biological replicates). The expression for each gene was normalized against Actb, and the relative level between samples was calculated using the 2^(-∆∆Ct) method. The bar charts show the mean ± s.e.m. P values are from two-sided t-test. c, Flow cytometry analysis for EPCAM+ and EPCAM− populations in KPFCT/Grem1wt/wt and KPFCT/Grem1Δ/Δ organoids. d, Principal component analysis (PCA) plot for RNA-seq data from KPFCT/Grem1wt/wt and KPFCT/Grem1Δ/Δ tumours (n = 5 mice). e, Nearest Template Prediction (NTP) subtype correlation analysis on KPFCT/Grem1wt/wt and KPFCT/Grem1Δ/Δ tumours using RNA-seq data (n = 5 mice). Collison et al’s PDAssigner was used for classical and quasi-mesenchymal subtype prediction. f, GSEA plots for enrichment of the classical or quasi-mesenchymal subtype for KPFCT/Grem1wt/wt and KPFCT/Grem1Δ/Δ tumours. FDR, false discovery rate.
Extended Data Fig. 6 Grem1 deletion promotes liver metastasis of pancreatic cancer.
a, b, Schemes depicting the KPhetFC/Grem1fl/fl mouse model and the experimental approach. * denotes the G12D mutation site in the KrasFSF-G12D allele. Yellow arrowhead, loxP sites; green arrowhead, Frt sites; TMX, tamoxifen. c, Images for liver and diaphragm tissues from KPhetFC/Grem1wt/wt and KPhetFC/Grem1Δ/Δ mice. Red arrowheads indicate metastatic foci in the liver; the black arrowhead indicates metastatic foci in the diaphragm. d, Percentage of mice developing liver metastases in KPhetFC/Grem1wt/wt (n = 6) and KPhetFC/Grem1Δ/Δ (n = 8) mice. The P values are shown above the chart; two-tailed Fisher's exact test. e, Quantification of the number of liver metastases per mouse in KPhetFC/Grem1wt/wt (n = 6) and KPhetFC/Grem1Δ/Δ (n = 8) mice. The scatter dot charts show the mean ± s.e.m. P value is from two-sided Mann-Whitney test. f, H&E and IHC staining for CK19 in liver metastases from KPhetFC/Grem1wt/wt and KPhetFC/Grem1Δ/Δ mice (n = 2 for KPhetFC/Grem1wt/wt and n = 5 for KPhetFC/Grem1Δ/Δ). The dashed circles/lines indicate metastases. Scale bar, 200 μm. g, Images for lung tissues from KPhetFC/Grem1wt/wt and KPhetFC/Grem1Δ/Δ mice. Red arrowheads indicate metastatic foci in the lung. h, Percentage of mice developing lung metastases in KPhetFC/Grem1wt/wt (n = 6) and KPhetFC/Grem1Δ/Δ (n = 7) mice. i, Quantification of the number of lung metastases per mouse in KPhetFC/Grem1wt/wt (n = 6) and KPhetFC/Grem1Δ/Δ (n = 7) mice. The scatter dot charts show the mean ± s.e.m. j, H&E and IHC staining for CK19 in lung metastases from KPhetFC/Grem1wt/wt and KPhetFC/Grem1Δ/Δ mice (n = 5). The dashed circles indicate metastases. Scale bar, 400 μm. k, GREM1 expression in normal tissues, pancreatic tumours and metastases from a microarray dataset. The scatter charts show the mean ± s.e.m. P values are from two-sided Mann-Whitney test.
Extended Data Fig. 7 Grem1 deletion reduces Grem1-expressing myCAFs but does not affect iCAFs, apCAFs and infiltrated immune cells.
a, FACS plots for the separation of epithelial tumour cells (ETCs), mesenchymal tumour cells (MTCs) and CAFs from KPCY tumours using YFP, EPCAM and PDPN markers, which mark tumour cells, epithelial cells and CAFs, respectively. Str., stroma; Tu, Tumour. b, RT–qPCR analysis of Grem1 on FACS-sorted ETCs, MTCs and CAFs cells from KPCY tumours (n = 4 mice). c, FACS plots for the separation of myCAFs and iCAFs from KPCY tumours using PDPN and Ly6C markers. Stromal cells were distinguished from tumour cells by YFP, and PDPN+Ly6C− and PDPN+Ly6C− label myCAFs and iCAFs in the stroma, respectively. Str., stroma; Tu, Tumour. d, RT–qPCR analysis of Grem1 on FACS-sorted ETCs, MTCs and CAFs cells from KPCY tumour (n = 3 mice). e, Immunofluorescent staining of aSMA, vimentin and GFP in KPFCT/Grem1wt/wt and KPFCT/Grem1Δ/Δ tumours. The dashed circle indicates a highly mesenchymal tumour area with fewer aSMA+ CAFs. Scale bar, 400 μm. f, Quantification of aSMA+ areas in KPFCT/Grem1wt/wt and KPFCT/Grem1Δ/Δ tumours (n = 4 mice). For each tumour, four representative regions with the area of 2.35 mm3 were counted and averaged. g, GSEA plots for the gene signatures of myCAFs, apCAFs and iCAFs in KPFCT/Grem1wt/wt and KPFCT/Grem1Δ/Δ tumours (n = 5 mice). h, Quantification of different types of infiltrated immune cells in KPFCT/Grem1wt/wt and KPFCT/Grem1Δ/Δ tumours by flow cytometry (n = 3 mice). For b, d, gene expression was normalized to Rplp0, and the relative level between samples was calculated using the 2^(-∆∆Ct) method. The bar charts show the mean ± s.e.m. P values are from one-way ANOVA and Tukey multiple comparison test (b), two-sided t-test (d, h), and two-sided Mann-Whitney test (f); NS, not significant.
Extended Data Fig. 8 Grem1 overexpression induces epithelial differentiation of pancreatic cancer.
a, RT–qPCR analysis of Grem1 on KPFCT organoids infected with control (pLV) or Grem1-overexpressing (pLV-Grem1) lentiviruses (n = 3 biological replicates). b, Flow cytometry analysis of the EPCAM− population in KPFCT organoids infected with control or Grem1-overexpressing lentiviruses, treated with vehicle or 100 ng/mL BMP2. c, Immunofluorescent staining of GFP, CK19 and vimentin on subcutaneous tumour grafts derived from KPFCT organoids infected with control or Grem1-overexpressing lentiviruses (n = 3 mice). Scale bar, 1000 μm. Indicated areas are magnified on the bottom. Scale bar, 100 μm. d, Quantification of the number of CK19+vimentin−, CK19+vimentin− and CK19+vimentin− tumour cells in subcutaneous grafts by KPFCT organoids infected with control or Grem1-overexpressing lentiviruses (n = 3 mice). e, RT–qPCR analysis of GREM1 on PANC1 cells infected with control or GREM1-overexpressing lentiviruses (n = 3 biological replicates). f, RT–qPCR analysis of epithelial and mesenchymal markers on PANC1 cells infected with control or GREM1-overexpressing lentiviruses, treated with vehicle or 100 ng/mL BMP2 (n = 3 biological replicates). For a, e and f, gene expression was normalized against Actb, and the relative level between samples was calculated using the 2^(-∆∆Ct) method. The bar charts show the mean ± s.e.m. P values are from two-sided t-test (a, d, e) and one-way ANOVA and Tukey multiple comparison test (f).
Extended Data Fig. 9 Time-course analysis of EMT post Grem1 deletion in KPFC tumour grafts and organoids.
a, RT–qPCR analysis of Grem1 on FACS-sorted RFP+ cells from KPF/Grem1fl/fl tumour grafts at day 1 post vehicle or TMX treatment (n = 3 mice). The expression was normalized to Rplp0, and the relative level between samples was calculated using the 2^(-∆∆Ct) method. The bar charts show the mean ± s.e.m. b–d, Representative images for H&E and IHC staining for CK19 in KPFC/Grem1fl/fl tumour grafts at different time points post vehicle or TMX treatment (n = 3 mice). Scale bar, 500 μm. Indicated areas are magnified on the right. Scale bar, 100 μm. e, Scheme depicting the experimental approach for time-course analysis of EMT in organoids. f, Immunofluorescent staining of E-cad and vimentin in KPFC/Grem1fl/fl organoids at different time points of vehicle or 4-OHT treatment (n = 4 biological replicates). Organoids were switched to DMEM medium with 5% FCS before adding vehicle or 4-OHT. Scale bar, 20 μm. g, Quantification of vimentin+ cells in organoids in conditions shown in f (n = 4 biological replicates). The bar charts show the mean ± s.e.m. P values are from two-sided t-test.
Extended Data Fig. 10 Grem1 mediates a negative feedback mechanism for BMP2 signalling to inhibit EMT.
a, GSEA plot for enrichment of the gene ontology (GO) term SMAD binding in KPCY EPCAM− tumour cells. NES, normalized enrichment score. b, Heatmap for the KEGG TGF-beta pathway enriched in GREM1hi PDACs from the TCGA dataset. c, GSEA plot for enrichment of the KEGG TGF-beta pathway in GREM1hi PDACs from the TCGA dataset. NES, normalized enrichment score. FDR, false discovery rate. d, RT–qPCR analysis of BMP receptor genes on sorted KPCY EPCAM+ and EPCAM− tumour cells (n = 3 mice). The expression was normalized to Rplp0, and the relative level between samples was calculated using the 2^(-∆∆Ct) method. The bar charts show the mean ± s.e.m. e, Western blotting of BMPR2, BMPR1A, EPCAM and β-actin on sorted EPCAM+ and EPCAM− cells from KPFC/Grem1fl/fl organoids (n = 3 independent experiments). The numbers indicate the molecular weight (kDa). For gel source data, see Supplementary Figure 1. f, g, KEGG TGF-β signalling and SMAD binding score plots for GREM1pos and GREM1neg cancer cells identified in a human PDAC scRNA-seq dataset. P values are from two-sided Wilcoxon test. h, Dot plot for the expression of BMP related genes in GREM1pos and GREM1neg cancer cells identified in a human PDAC scRNA-seq dataset. i, j, SMAD5 binding motifs and scores in Grem1 promoter, predicted by the JASAPR database. k, Immunofluorescent staining of E-cad and vimentin in control (pLV) or Grem1-overexpressing (pLV-Grem1) KPFC/Grem1fl/fl organoids, treated as indicated (n = 3 biological replicates). Scale bar, 100 μm. l, Quantification of the number of vimentin+ organoids in conditions shown in k (n = 3 biological replicates). The bar charts show the mean ± s.e.m. P values are from two-way ANOVA and Tukey multiple comparison test.
Extended Data Fig. 11 Upregulation of core EMT transcription factors in mesenchymal PDAC cancer cells and their activation by BMP2 or Grem1 deletion.
a, Fold changes of mRNA expression of EMT-TFs between EPCAM+ and EPCAM− tumour cells from fresh KPCY tumours (n = 3 mice), quantified by RT–qPCR. b, RT–qPCR analysis of Snail, Slug, Zeb1, and Twist on sorted EPCAM+ and EPCAM− populations from KPFC organoids, treated as indicated (n = 3 mice). c, RT–qPCR analysis of EMT TF genes on KPFCT/Grem1wt/wt and KPFCT/Grem1Δ/Δ organoids (n = 3 biological replicates). d, Immunofluorescence of Zeb1, GFP and vimentin in primary KPFCT/Grem1wt/wt and KPFCT/Grem1Δ/Δ tumours and KPhetFC/Grem1wt/wt and KPhetFC/Grem1Δ/Δ metastases (n = 3 mice). Scale bar, 100 μm. e. SMAD5 binding motifs and scores in Snail promoter, predicted by the JASAPR database. f, ChIP analysis of pSMAD1/5/9 binding sites on Snail promoter in KPFC/Grem1fl/fl cells, treated as indicated. IgG control or phospho-SMAD1Ser463/465/SMAD5Ser463/465/SMAD9Ser465/467 antibodies were used for ChIP and DNA was quantified by qPCR. DNA levels for each condition were normalized to the input, and the fold-change was calculated over the vehicle control (n = 3 independent experiments). For a, gene expression was normalized to Rplp0, and for b and c, gene expression was normalized to Actb; the relative level between samples was calculated using the 2^(-∆∆Ct) method. The bar charts show the mean ± s.e.m. P values are from one-way ANOVA and Tukey multiple comparison test (b, f) and two-sided t-test (c).
Extended Data Fig. 12 Snail and Slug are essential TFs that act together downstream of Grem1 to regulate EMT.
a, RT–qPCR analysis of Snail and Slug on shControl and Snail and Slug dual knocked-down (shSnail/Slug_comb1 and shSnail/Slug_comb2) KPFC/Grem1fl/fl organoids (n = 3 biological replicates). b, RT–qPCR analysis of Vim and Zeb1 in shControl shSnail/Slug_comb1, shSnail/Slug_comb2 and Snail knocked-down (shSnail_1) KPFC/Grem1fl/fl organoids (n = 3 biological replicates). c, Immunofluorescent staining of vimentin in shControl shSnail/Slug_comb1, shSnail/Slug_comb2 and shSnail_1 KPFC/Grem1fl/fl organoids, treated as indicated (n = 3 biological replicates). Scale bar, 50 μm. d, Quantification of the number of vimentin+ cells in organoids in conditions shown in c (n = 3 biological replicates). e, Representative images for H&E and IHC staining for CK19 in shControl and shSnail/Slug tumour grafts, treated with vehicle or TMX (n = 3 mice). Scale bar, 250 μm. For a and b, the expression for each gene was normalized to Actb; the relative level between samples was calculated using the 2^(-∆∆Ct) method. The bar charts show the mean ± s.e.m. P values are from two-sided t-test (a) and one-way ANOVA and Tukey multiple comparison test (b, d).
Extended Data Fig. 13 Model for the mechanism of Grem1 in maintaining cellular heterogeneity in pancreatic cancer.
a, Schematic cartoon depicting the mechanism for the Grem1-mediated paracrine interaction between epithelial and mesenchymal cancer cells, which maintains the epithelial fate by restricting BMP signalling and EMT. b, The BMP2-Grem1 self-inhibitory feedback loop that mimics the Turing’s reaction-diffusion model.
Supplementary information
Supplementary Information
Supplementary Figs. 1 and 2 and Supplementary Tables 1–4.
Source data
Rights and permissions
About this article
Cite this article
Lan, L., Evan, T., Li, H. et al. GREM1 is required to maintain cellular heterogeneity in pancreatic cancer. Nature 607, 163–168 (2022). https://doi.org/10.1038/s41586-022-04888-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-022-04888-7
This article is cited by
-
PRRX1-TOP2A interaction is a malignancy-promoting factor in human malignant peripheral nerve sheath tumours
British Journal of Cancer (2024)
-
Molecular profile of metastasis, cell plasticity and EMT in pancreatic cancer: a pre-clinical connection to aggressiveness and drug resistance
Cancer and Metastasis Reviews (2024)
-
The epithelial–mesenchymal plasticity landscape: principles of design and mechanisms of regulation
Nature Reviews Genetics (2023)
-
The role of epithelial-mesenchymal transition and autophagy in pancreatic ductal adenocarcinoma invasion
Cell Death & Disease (2023)
-
Evaluation of GREM1 and THBS2 as prognostic markers in in non-small cell lung cancer
Journal of Cancer Research and Clinical Oncology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.