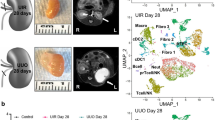
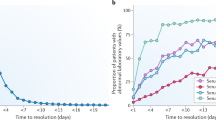
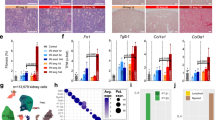
Abstract
The kidney epithelium, with its intricate arrangement of highly specialized cell types, constitutes the functional core of the organ. Loss of kidney epithelium is linked to the loss of functional nephrons and a subsequent decline in kidney function. In kidney transplantation, epithelial injury signatures observed during post-transplantation surveillance are strong predictors of adverse kidney allograft outcomes. However, epithelial injury is currently neither monitored clinically nor addressed therapeutically after kidney transplantation. Several factors can contribute to allograft epithelial injury, including allograft rejection, drug toxicity, recurrent infections and postrenal obstruction. The injury mechanisms that underlie allograft injury overlap partially with those associated with acute kidney injury (AKI) and chronic kidney disease (CKD) in the native kidney. Studies using advanced transcriptomic analyses of single cells from kidney or urine have identified a role for kidney injury-induced epithelial cell states in exacerbating and sustaining damage in AKI and CKD. These epithelial cell states and their associated expression signatures are also observed in transplanted kidney allografts, suggesting that the identification and characterization of transcriptomic epithelial cell states in kidney allografts may have potential clinical implications for diagnosis and therapy.
Key points
-
Transcriptomic studies using single-cell sequencing and spatial transcriptomics have identified epithelial cell populations associated with acute kidney injury (AKI) and chronic kidney disease (CKD) in mouse and human kidneys.
-
These injured epithelial cell populations are especially predominant in the proximal tubules but extend to other cell types, particularly in human AKI and CKD.
-
AKI- and CKD-associated epithelial cell populations show a marked downregulation of marker genes associated with the healthy kidney epithelium and can show a pro-inflammatory and pro-fibrotic gene expression profile.
-
Some injured epithelial cell populations can persist for a period of time after AKI and in CKD, attracting and activating fibroblasts and leukocytes, and thereby potentially promoting further kidney damage.
-
Epithelial injury is an under-appreciated major risk factor of allograft survival after episodes of rejection in kidney transplants.
-
Molecular signatures of injured epithelial cell populations in kidney transplants after post-transplantation complications are highly reminiscent of those seen in AKI and CKD, and offer potential new avenues for monitoring and therapy.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Abecassis, M. et al. Kidney transplantation as primary therapy for end-stage renal disease: a National Kidney Foundation/Kidney Disease Outcomes Quality Initiative (NKF/KDOQITM) conference. Clin. J. Am. Soc. Nephrol. 3, 471–480 (2008).
Lentine, K. L. et al. OPTN/SRTR 2021 annual data report: kidney. Am. J. Transpl. 23, S21–S120 (2023).
Halloran, P. F., Famulski, K. S. & Reeve, J. Molecular assessment of disease states in kidney transplant biopsy samples. Nat. Rev. Nephrol. 12, 534–548 (2016).
Madill-Thomsen, K. S. et al. Relating molecular T cell-mediated rejection activity in kidney transplant biopsies to time and to histologic tubulitis and atrophy-fibrosis. Transplantation 107, 1102–1114 (2023).
Halloran, P. F. et al. Review: the transcripts associated with organ allograft rejection. Am. J. Transpl. 18, 785–795 (2018).
Liu, Y. et al. Single-cell analysis reveals immune landscape in kidneys of patients with chronic transplant rejection. Theranostics 10, 8851–8862 (2020).
Malone, A. F. et al. Harnessing expressed single nucleotide variation and single cell RNA sequencing to define immune cell chimerism in the rejecting kidney transplant. J. Am. Soc. Nephrol. 31, 1977–1986 (2020).
Rashmi, P. et al. Multiplexed droplet single-cell sequencing (Mux-Seq) of normal and transplant kidney. Am. J. Transpl. 22, 876–885 (2022).
Wu, H. et al. Single-cell transcriptomics of a human kidney allograft biopsy specimen defines a diverse inflammatory response. J. Am. Soc. Nephrol. 29, 2069–2080 (2018).
McDaniels, J. M. et al. Single nuclei transcriptomics delineates complex immune and kidney cell interactions contributing to kidney allograft fibrosis. Kidney Int. 103, 1077–1092 (2023).
Suryawanshi, H. et al. Detection of infiltrating fibroblasts by single-cell transcriptomics in human kidney allografts. PLoS ONE 17, e0267704 (2022).
Kellum, J. A. & Lameire, N. Diagnosis, evaluation, and management of acute kidney injury: a KDIGO summary (Part 1). Crit. Care 17, 204 (2013).
Hoste, E. A. J. et al. Global epidemiology and outcomes of acute kidney injury. Nat. Rev. Nephrol. 14, 607–625 (2018).
Vijayan, A. Tackling AKI: prevention, timing of dialysis and follow-up. Nat. Rev. Nephrol. 17, 87–88 (2021).
Ronco, C. Acute kidney injury: from clinical to molecular diagnosis. Crit. Care 20, 201 (2016).
Kane-Gill, S. L. et al. Risk factors for acute kidney injury in older adults with critical illness: a retrospective cohort study. Am. J. Kidney Dis. 65, 860–869 (2015).
Coca, S. G., Yusuf, B., Shlipak, M. G., Garg, A. X. & Parikh, C. R. Long-term risk of mortality and other adverse outcomes after acute kidney injury: a systematic review and meta-analysis. Am. J. Kidney Dis. 53, 961–973 (2009).
Newsome, B. B. et al. Long-term risk of mortality and end-stage renal disease among the elderly after small increases in serum creatinine level during hospitalization for acute myocardial infarction. Arch. Intern. Med. 168, 609–616 (2008).
Abdala, P. M., Swanson, E. A. & Hutchens, M. P. Meta-analysis of AKI to CKD transition in perioperative patients. Perioper. Med. 10, 24 (2021).
Guzzi, F., Cirillo, L., Roperto, R. M., Romagnani, P. & Lazzeri, E. Molecular mechanisms of the acute kidney injury to chronic kidney disease transition: an updated view. Int. J. Mol. Sci. 20, 4941 (2019).
Fogo, A. B., Lusco, M. A., Najafian, B. & Alpers, C. E. AJKD atlas of renal pathology: ischemic acute tubular injury. Am. J. Kidney Dis. 67, e25 (2016).
Fogo, A. B., Lusco, M. A., Najafian, B. & Alpers, C. E. AJKD atlas of renal pathology: toxic acute tubular injury. Am. J. Kidney Dis. 67, e31–e32 (2016).
Liu, J. et al. Molecular characterization of the transition from acute to chronic kidney injury following ischemia/reperfusion. JCI Insight 2, e94716 (2017).
Cippa, P. E. & McMahon, A. P. Proximal tubule responses to injury: interrogation by single-cell transcriptomics. Curr. Opin. Nephrol. Hypertens. 32, 352–358 (2023).
Xu, K. et al. Unique transcriptional programs identify subtypes of AKI. J. Am. Soc. Nephrol. 28, 1729–1740 (2017).
Vigolo, E. et al. Canonical BMP signaling in tubular cells mediates recovery after acute kidney injury. Kidney Int. 95, 108–122 (2019).
Marko, L. et al. Tubular epithelial NF-κB activity regulates ischemic AKI. J. Am. Soc. Nephrol. 27, 2658–2669 (2016).
Supavekin, S. et al. Differential gene expression following early renal ischemia/reperfusion. Kidney Int. 63, 1714–1724 (2003).
Yuen, P. S., Jo, S. K., Holly, M. K., Hu, X. & Star, R. A. Ischemic and nephrotoxic acute renal failure are distinguished by their broad transcriptomic responses. Physiol. Genomics 25, 375–386 (2006).
Yoshida, T. et al. Monitoring changes in gene expression in renal ischemia-reperfusion in the rat. Kidney Int. 61, 1646–1654 (2002).
Humphreys, B. D. et al. Intrinsic epithelial cells repair the kidney after injury. Cell Stem Cell 2, 284–291 (2008).
Gerhardt, L. M. S. & McMahon, A. P. Identifying common molecular mechanisms in experimental and human acute kidney injury. Semin. Nephrol. 42, 151286 (2022).
Chang-Panesso, M. & Humphreys, B. D. Cellular plasticity in kidney injury and repair. Nat. Rev. Nephrol. 13, 39–46 (2017).
Chang-Panesso, M. et al. FOXM1 drives proximal tubule proliferation during repair from acute ischemic kidney injury. J. Clin. Invest. 129, 5501–5517 (2019).
Humphreys, B. D. Kidney injury, stem cells and regeneration. Curr. Opin. Nephrol. Hypertens. 23, 25–31 (2014).
Lindgren, D. et al. Isolation and characterization of progenitor-like cells from human renal proximal tubules. Am. J. Pathol. 178, 828–837 (2011).
Angelotti, M. L. et al. Characterization of renal progenitors committed toward tubular lineage and their regenerative potential in renal tubular injury. Stem Cell 30, 1714–1725 (2012).
Loverre, A. et al. Increase of proliferating renal progenitor cells in acute tubular necrosis underlying delayed graft function. Transplantation 85, 1112–1119 (2008).
Peired, A. J. et al. Acute kidney injury promotes development of papillary renal cell adenoma and carcinoma from renal progenitor cells. Sci. Transl. Med. 12, eaaw6003 (2020).
Scholz, H. et al. Kidney physiology and susceptibility to acute kidney injury: implications for renoprotection. Nat. Rev. Nephrol. 17, 335–349 (2021).
Shen, T. H. et al. Snapshots of nascent RNA reveal cell- and stimulus-specific responses to acute kidney injury. JCI Insight 7, e146374 (2022).
Liu, J. et al. Cell-specific translational profiling in acute kidney injury. J. Clin. Invest. 124, 1242–1254 (2014).
Doyle, J. P. et al. Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell 135, 749–762 (2008).
Heiman, M. et al. A translational profiling approach for the molecular characterization of CNS cell types. Cell 135, 738–748 (2008).
Kirita, Y., Wu, H., Uchimura, K., Wilson, P. C. & Humphreys, B. D. Cell profiling of mouse acute kidney injury reveals conserved cellular responses to injury. Proc. Natl Acad. Sci. USA 117, 15874–15883 (2020).
Gerhardt, L. M. S. et al. Lineage tracing and single-nucleus multiomics reveal novel features of adaptive and maladaptive repair after acute kidney injury. J. Am. Soc. Nephrol. 34, 554–571 (2023).
Dixon, E. E., Wu, H., Muto, Y., Wilson, P. C. & Humphreys, B. D. Spatially resolved transcriptomic analysis of acute kidney injury in a female murine model. J. Am. Soc. Nephrol. 33, 279–289 (2022).
Hinze, C. et al. Single-cell transcriptomics reveals common epithelial response patterns in human acute kidney injury. Genome Med. 14, 103 (2022).
Klocke, J. et al. Urinary single-cell sequencing captures kidney injury and repair processes in human acute kidney injury. Kidney Int. 102, 1359–1370 (2022).
Rudman-Melnick, V. et al. Single-cell profiling of AKI in a murine model reveals novel transcriptional signatures, profibrotic phenotype, and epithelial-to-stromal crosstalk. J. Am. Soc. Nephrol. 31, 2793–2814 (2020).
Balzer, M. S. et al. Single-cell analysis highlights differences in druggable pathways underlying adaptive or fibrotic kidney regeneration. Nat. Commun. 13, 4018 (2022).
Legouis, D. et al. Single cell profiling in COVID-19 associated acute kidney injury reveals patterns of tubule injury and repair in human. Preprint at bioRxiv https://doi.org/10.1101/2021.10.05.463150 (2021).
Jansen, J. et al. SARS-CoV-2 infects the human kidney and drives fibrosis in kidney organoids. Cell Stem Cell 29, 217–231.e8 (2022).
Kuppe, C. et al. Decoding myofibroblast origins in human kidney fibrosis. Nature 589, 281–286 (2021).
Lake, B. B. et al. An atlas of healthy and injured cell states and niches in the human kidney. Nature 619, 585–594 (2023).
Gerhardt, L. M. S., Liu, J., Koppitch, K., Cippa, P. E. & McMahon, A. P. Single-nuclear transcriptomics reveals diversity of proximal tubule cell states in a dynamic response to acute kidney injury. Proc. Natl Acad. Sci. USA 118, e2026684118 (2021).
Yang, L., Besschetnova, T. Y., Brooks, C. R., Shah, J. V. & Bonventre, J. V. Epithelial cell cycle arrest in G2/M mediates kidney fibrosis after injury. Nat. Med. 16, 535–543 (2010).
Friedmann Angeli, J. P. et al. Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice. Nat. Cell Biol. 16, 1180–1191 (2014).
Ide, S. et al. Ferroptotic stress promotes the accumulation of pro-inflammatory proximal tubular cells in maladaptive renal repair. Elife 10, e68603 (2021).
Linkermann, A. et al. Synchronized renal tubular cell death involves ferroptosis. Proc. Natl Acad. Sci. USA 111, 16836–16841 (2014).
Zhao, Z. et al. XJB-5-131 inhibited ferroptosis in tubular epithelial cells after ischemia-reperfusion injury. Cell Death Dis. 11, 629 (2020).
Li, H., Dixon, E. E., Wu, H. & Humphreys, B. D. Comprehensive single-cell transcriptional profiling defines shared and unique epithelial injury responses during kidney fibrosis. Cell Metab. 34, 1977–1998.e9 (2022).
Chen, Z. et al. Single-cell sequencing reveals homogeneity and heterogeneity of the cytopathological mechanisms in different etiology-induced AKI. Cell Death Dis. 14, 318 (2023).
Liu, Y. Cellular and molecular mechanisms of renal fibrosis. Nat. Rev. Nephrol. 7, 684–696 (2011).
Humphreys, B. D. Mechanisms of renal fibrosis. Annu. Rev. Physiol. 80, 309–326 (2018).
Li, L., Fu, H. & Liu, Y. The fibrogenic niche in kidney fibrosis: components and mechanisms. Nat. Rev. Nephrol. 18, 545–557 (2022).
Tapmeier, T. T. et al. Pivotal role of CD4+ T cells in renal fibrosis following ureteric obstruction. Kidney Int. 78, 351–362 (2010).
Anders, H. J. et al. A chemokine receptor CCR-1 antagonist reduces renal fibrosis after unilateral ureter ligation. J. Clin. Invest. 109, 251–259 (2002).
Gewin, L. The many talents of transforming growth factor-β in the kidney. Curr. Opin. Nephrol. Hypertens. 28, 203–210 (2019).
Floege, J., Eitner, F. & Alpers, C. E. A new look at platelet-derived growth factor in renal disease. J. Am. Soc. Nephrol. 19, 12–23 (2008).
Livingston, M. J. et al. Tubular cells produce FGF2 via autophagy after acute kidney injury leading to fibroblast activation and renal fibrosis. Autophagy 19, 256–277 (2023).
Zhu, H. et al. Tenascin-C promotes acute kidney injury to chronic kidney disease progression by impairing tubular integrity via ɑvβ6 integrin signaling. Kidney Int. 97, 1017–1031 (2020).
Abedini, A. et al. Spatially resolved human kidney multi-omics single cell atlas highlights the key role of the fibrotic microenvironment in kidney disease progression. Preprint at bioRxiv https://doi.org/10.1101/2022.10.24.513598 (2022).
Nagao, T. et al. Osteopontin plays a critical role in interstitial fibrosis but not glomerular sclerosis in diabetic nephropathy. Nephron Extra 2, 87–103 (2012).
Sato, Y. & Yanagita, M. Resident fibroblasts in the kidney: a major driver of fibrosis and inflammation. Inflamm. Regen. 37, 17 (2017).
Coca, S. G. Acute kidney injury in elderly persons. Am. J. Kidney Dis. 56, 122–131 (2010).
Stevens, L. A., Viswanathan, G. & Weiner, D. E. Chronic kidney disease and end-stage renal disease in the elderly population: current prevalence, future projections, and clinical significance. Adv. Chronic Kidney Dis. 17, 293–301 (2010).
Tonelli, M. et al. Systematic review: kidney transplantation compared with dialysis in clinically relevant outcomes. Am. J. Transpl. 11, 2093–2109 (2011).
Bastani, B. The present and future of transplant organ shortage: some potential remedies. J. Nephrol. 33, 277–288 (2020).
Ponticelli, C., Villa, M., Cesana, B., Montagnino, G. & Tarantino, A. Risk factors for late kidney allograft failure. Kidney Int. 62, 1848–1854 (2002).
Kwon, H. et al. Pure T-cell mediated rejection following kidney transplant according to response to treatment. PLoS ONE 16, e0256898 (2021).
Kant, S., Dasgupta, A., Bagnasco, S. & Brennan, D. C. BK virus nephropathy in kidney transplantation: a state-of-the-art review. Viruses 14, 1616 (2022).
Requiao-Moura, L. R., Durão Junior, M. de S., Matos, A. C. & Pacheco-Silva, A. Ischemia and reperfusion injury in renal transplantation: hemodynamic and immunological paradigms. Einstein 13, 129–135 (2015).
Mbianda, C., El-Meanawy, A. & Sorokin, A. Mechanisms of BK virus infection of renal cells and therapeutic implications. J. Clin. Virol. 71, 59–62 (2015).
Halloran, P. F. et al. Molecular diagnosis of ABMR with or without donor-specific antibody in kidney transplant biopsies: differences in timing and intensity but similar mechanisms and outcomes. Am. J. Transpl. 22, 1976–1991 (2022).
Cippa, P. E. et al. Transcriptional trajectories of human kidney injury progression. JCI Insight 3, e123151 (2018).
Famulski, K. S. et al. Molecular phenotypes of acute kidney injury in kidney transplants. J. Am. Soc. Nephrol. 23, 948–958 (2012).
O’Connell, P. J. et al. Biopsy transcriptome expression profiling to identify kidney transplants at risk of chronic injury: a multicentre, prospective study. Lancet 388, 983–993 (2016).
Ke, B., Fan, C., Yang, L. & Fang, X. Matrix metalloproteinases-7 and kidney fibrosis. Front. Physiol. 8, 21 (2017).
Nankivell, B. J. et al. The natural history of chronic allograft nephropathy. N. Engl. J. Med. 349, 2326–2333 (2003).
Loupy, A. et al. The Banff 2019 Kidney Meeting report (I): updates on and clarification of criteria for T cell- and antibody-mediated rejection. Am. J. Transpl. 20, 2318–2331 (2020).
Jeong, H. J. Diagnosis of renal transplant rejection: Banff classification and beyond. Kidney Res. Clin. Pract. 39, 17–31 (2020).
Chandran, S. & Mannon, R. B. T cell-mediated rejection in kidney transplant recipients: the end(point) is also the beginning. Am. J. Transpl. 22, 683–684 (2022).
Bohmig, G. A., Eskandary, F., Doberer, K. & Halloran, P. F. The therapeutic challenge of late antibody-mediated kidney allograft rejection. Transpl. Int. 32, 775–788 (2019).
Ho, J. et al. Effectiveness of T cell-mediated rejection therapy: a systematic review and meta-analysis. Am. J. Transpl. 22, 772–785 (2022).
Townamchai, N. & Avihingsanon, Y. Updated management for antibody-mediated rejection: opportunity to prolong kidney allograft survival. Curr. Opin. Nephrol. Hypertens. 32, 13–19 (2023).
Einecke, G. et al. Factors associated with kidney graft survival in pure antibody-mediated rejection at the time of indication biopsy: importance of parenchymal injury but not disease activity. Am. J. Transpl. 21, 1391–1401 (2021).
Halloran, P. F. et al. Discovering novel injury features in kidney transplant biopsies associated with TCMR and donor aging. Am. J. Transpl. 21, 1725–1739 (2021).
Halloran, P. F. et al. Archetypal analysis of injury in kidney transplant biopsies identifies two classes of early AKI. Front. Med. 9, 817324 (2022).
Lamarthee, B. et al. Transcriptional and spatial profiling of the kidney allograft unravels a central role for FcyRIII+ innate immune cells in rejection. Nat. Commun. 14, 4359 (2023).
Wang, Y. et al. Single-cell RNA-seq analysis identified kidney progenitor cells from human urine. Protein Cell 12, 305–312 (2021).
Abedini, A. et al. Urinary single-cell profiling captures the cellular diversity of the kidney. J. Am. Soc. Nephrol. 32, 614–627 (2021).
Latt, K. Z. et al. Urine single-cell RNA sequencing in focal segmental glomerulosclerosis reveals inflammatory signatures. Kidney Int. Rep. 7, 289–304 (2022).
Cheung, M. D. et al. Single-cell RNA sequencing of urinary cells reveals distinct cellular diversity in COVID-19-associated AKI. Kidney360 3, 28–36 (2022).
Goerlich, N. et al. Kidney transplant monitoring by urinary flow cytometry: biomarker combination of T cells, renal tubular epithelial cells, and podocalyxin-positive cells detects rejection. Sci. Rep. 10, 796 (2020).
Galante, N. Z. et al. Noninvasive immune monitoring assessed by flow cytometry and real time RT-PCR in urine of renal transplantation recipients. Transpl. Immunol. 16, 73–80 (2006).
Doke, T. et al. Single-cell analysis identifies the interaction of altered renal tubules with basophils orchestrating kidney fibrosis. Nat. Immunol. 23, 947–959 (2022).
Amor, C. et al. Senolytic CAR T cells reverse senescence-associated pathologies. Nature 583, 127–132 (2020).
Mylonas, K. J. et al. Cellular senescence inhibits renal regeneration after injury in mice, with senolytic treatment promoting repair. Sci. Transl. Med. 13, eabb0203 (2021).
Dhillon, P. et al. The nuclear receptor ESRRA protects from kidney disease by coupling metabolism and differentiation. Cell Metab. 33, 379–394.e8 (2021).
Taguchi, K. et al. Cyclin G1 induces maladaptive proximal tubule cell dedifferentiation and renal fibrosis through CDK5 activation. J. Clin. Invest. 132, e158096 (2022).
Author information
Authors and Affiliations
Contributions
All authors researched data for the article, made substantial contributions to discussions of the content and wrote, reviewed, and/or edited the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Nephrology thanks Benjamin Humphreys, Maarten Naesens and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hinze, C., Lovric, S., Halloran, P.F. et al. Epithelial cell states associated with kidney and allograft injury. Nat Rev Nephrol (2024). https://doi.org/10.1038/s41581-024-00834-0
Accepted:
Published:
DOI: https://doi.org/10.1038/s41581-024-00834-0