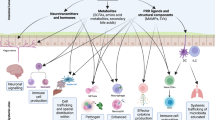
Abstract
Inflammatory bowel disease (IBD) is a chronic inflammatory condition of the gastrointestinal tract that results from dysfunction in innate and/or adaptive immune responses. Impaired innate immunity, which leads to lack of control of an altered intestinal microbiota and to activation of the adaptive immune system, promotes a secondary inflammatory response that is responsible for tissue damage. Neutrophils are key players in innate immunity in IBD, but their roles have been neglected compared with those of other immune cells. The latest studies on neutrophils in IBD have revealed unexpected complexities, with heterogeneous populations and dual functions, both deleterious and protective, for the host. In parallel, interconnections between disease development, intestinal microbiota and neutrophils have been highlighted. Numerous IBD susceptibility genes (such as NOD2, NCF4, LRRK2, CARD9) are involved in neutrophil functions related to defence against microorganisms. Moreover, severe monogenic diseases involving dysfunctional neutrophils, including chronic granulomatous disease, are characterized by intestinal inflammation that mimics IBD and by alterations in the intestinal microbiota. This observation demonstrates the dialogue between neutrophils, gut inflammation and the microbiota. Neutrophils affect microbiota composition and function in several ways. In return, microbial factors, including metabolites, regulate neutrophil production and function directly and indirectly. It is crucial to further investigate the diverse roles played by neutrophils in host–microbiota interactions, both at steady state and in inflammatory conditions, to develop new IBD therapies. In this Review, we discuss the roles of neutrophils in IBD, in light of emerging evidence proving strong interconnections between neutrophils and the gut microbiota, especially in an inflammatory context.
Key points
-
The study of neutrophils in inflammatory bowel disease (IBD) has revealed unexpected complexities, with heterogeneous populations and dual functions, both deleterious and protective, for the host.
-
Strong evidence supports an association between defects in neutrophil functions, alterations of the gut microbiota and intestinal inflammation.
-
Neutrophils influence microbiota composition and function, especially in inflammatory conditions.
-
The intestinal microbiota regulates neutrophil production and function, from activation to maturation.
-
Several microbiota-derived metabolites have been identified as modulators of neutrophil activity.
-
It is crucial to elucidate the roles played by neutrophils in host–microbiota interactions to develop new ways to tackle complex diseases associated with microbiota alterations, such as IBD.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Plichta, D. R., Graham, D. B., Subramanian, S. & Xavier, R. J. Therapeutic opportunities in inflammatory bowel disease: mechanistic dissection of host–microbiome relationships. Cell 178, 1041–1056 (2019).
Graham, D. B. & Xavier, R. J. Pathway paradigms revealed from the genetics of inflammatory bowel disease. Nature 578, 527–539 (2020).
Fernandes, F. P., Leal, V. N. C., Souza de Lima, D., Reis, E. C. & Pontillo, A. Inflammasome genetics and complex diseases: a comprehensive review. Eur. J. Hum. Genet. 28, 1307–1321 (2020).
Wéra, O., Lancellotti, P. & Oury, C. The dual role of neutrophils in inflammatory bowel diseases. J. Clin. Med. 5, 118 (2016).
Drury, B., Hardisty, G., Gray, R. D. & Ho, G. Neutrophil extracellular traps in inflammatory bowel disease: pathogenic mechanisms and clinical translation. Cell. Mol. Gastroenterol. Hepatol. 12, 321–333 (2021).
Hugot, J.-P. et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature 411, 599–603 (2001).
Ogura, Y. et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature 411, 603–606 (2001).
Deshmukh, H. S. et al. Critical role of NOD2 in regulating the immune response to Staphylococcus aureus. Infect. Immun. 77, 1376–1382 (2009).
Franke, A. et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat. Genet. 42, 1118–1125 (2010).
Wouters, C. H., Maes, A., Foley, K. P., Bertin, J. & Rose, C. D. Blau syndrome, the prototypic auto-inflammatory granulomatous disease. Pediatr. Rheumatol. Online J. 12, 33 (2014).
Magro, F. et al. European consensus on the histopathology of inflammatory bowel disease. J. Crohns Colitis 7, 827–851 (2013).
Lopez, R. N. et al. Fecal biomarkers in inflammatory bowel disease: faecal biomarkers. J. Gastroenterol. Hepatol. 32, 577–582 (2017).
Mortensen, J. H. et al. A specific calprotectin neo-epitope [CPa9-HNE] in serum from inflammatory bowel disease patients is associated with neutrophil activity and endoscopic severity. J. Crohns Colitis 16, 1447–1460 (2022).
Swaminathan, A. et al. Faecal myeloperoxidase as a biomarker of endoscopic activity in inflammatory bowel disease. J. Crohns Colitis 16, 1862–1873 (2022).
Mortha, A. et al. Neutralizing anti-granulocyte-macrophage–colony-stimulating factor autoantibodies recognize post-translational glycosylations on granulocyte-macrophage–colony-stimulating factor years before diagnosis and predict complicated Crohn’s disease. Gastroenterology 163, 659–670 (2022). This study shows that the detection of autoantibodies directed against GM-CSF could predict complicated ileal Crohn’s disease long before disease diagnosis.
Hamilton, J. A. GM-CSF in inflammation. J. Exp. Med. 217, e20190945 (2020).
Deniset, J. F. & Kubes, P. Neutrophil heterogeneity: bona fide subsets or polarization states? J. Leukoc. Biol. 103, 829–838 (2018).
Hidalgo, A. & Casanova-Acebes, M. Dimensions of neutrophil life and fate. Semin. Immunol. 57, 101506 (2021).
Garratt, L. W. Current understanding of the neutrophil transcriptome in health and disease. Cells 10, 2406 (2021).
Ng, L. G., Ostuni, R. & Hidalgo, A. Heterogeneity of neutrophils. Nat. Rev. Immunol. 19, 255–265 (2019).
Silvestre-Roig, C., Fridlender, Z. G., Glogauer, M. & Scapini, P. Neutrophil diversity in health and disease. Trends Immunol. 40, 565–583 (2019).
Burn, G. L., Foti, A., Marsman, G., Patel, D. F. & Zychlinsky, A. The neutrophil. Immunity 54, 1377–1391 (2021).
Zhang, D. & Frenette, P. S. Cross talk between neutrophils and the microbiota. Blood 133, 2168–2177 (2019).
Rosales, C. Neutrophil: a cell with many roles in inflammation or several cell types? Front. Physiol. 9, 113 (2018).
Hardisty, G. R. et al. High purity isolation of low density neutrophils casts doubt on their exceptionality in health and disease. Front. Immunol. 12, 625922 (2021).
Zhou, G. et al. CD177+ neutrophils as functionally activated neutrophils negatively regulate IBD. Gut 67, 1052–1063 (2018). This study identified a unique IBD-specific neutrophil population.
Hoogendijk, A. J. et al. Dynamic transcriptome–proteome correlation networks reveal human myeloid differentiation and neutrophil-specific programming. Cell Rep. 29, 2505–2519.e4 (2019). This study combines interesting high-throughput approaches to elucidate neutrophil biology.
Marini, O. et al. Mature CD10+ and immature CD10− neutrophils present in G-CSF-treated donors display opposite effects on T cells. Blood 129, 1343–1356 (2017).
Zhou, G. X. & Liu, Z. J. Potential roles of neutrophils in regulating intestinal mucosal inflammation of inflammatory bowel disease: role of neutrophils in IBD. J. Dig. Dis. 18, 495–503 (2017).
Buckley, C. D. et al. Identification of a phenotypically and functionally distinct population of long-lived neutrophils in a model of reverse endothelial migration. J. Leukoc. Biol. 79, 303–311 (2005).
Xu, Q., Zhao, W., Yan, M. & Mei, H. Neutrophil reverse migration. J. Inflamm. 19, 22 (2022).
Sullivan, D. P., Bui, T., Muller, W. A., Butin-Israeli, V. & Sumagin, R. In vivo imaging reveals unique neutrophil transendothelial migration patterns in inflamed intestines. Mucosal Immunol. 11, 1571–1581 (2018).
Xie, X. et al. Single-cell transcriptome profiling reveals neutrophil heterogeneity in homeostasis and infection. Nat. Immunol. 21, 1119–1133 (2020). This is an interesting study as it is one of the first to use single-cell transcriptomics to elucidate neutrophil biology at steady-state and during bacterial infection.
Wigerblad, G. et al. Single-cell analysis reveals the range of transcriptional states of circulating human neutrophils. J. Immunol. 209, 772–782 (2022).
Montaldo, E. et al. Cellular and transcriptional dynamics of human neutrophils at steady state and upon stress. Nat. Immunol. 23, 1470–1483 (2022).
Garrido-Trigo, A. et al. Macrophage neutrophil heterogeneity single-cell spatial resolution in hunam inflammatory bowel disease. Nat. Commun. 14, 4506 (2022).
Isles, H. M. et al. The CXCL12/CXCR4 signaling axis retains neutrophils at inflammatory sites in zebrafish. Front. Immunol. 10, 1784 (2019).
Ballesteros, I. et al. Co-option of neutrophil fates by tissue environments. Cell 183, 1282–1297.e18 (2020).
Hartl, D. et al. Infiltrated neutrophils acquire novel chemokine receptor expression and chemokine responsiveness in chronic inflammatory lung diseases. J. Immunol. 181, 8053–8067 (2008).
Chen, H., Wu, X., Xu, C., Lin, J. & Liu, Z. Dichotomous roles of neutrophils in modulating pathogenic and repair processes of inflammatory bowel diseases. Precis. Clin. Med. 4, 246–257 (2021).
Kang, L. et al. Neutrophil–epithelial crosstalk during intestinal inflammation. Cell. Mol. Gastroenterol. Hepatol. 14, 1257–1267 (2022).
Tarlton, J. F. et al. The role of up-regulated serine proteases and matrix metalloproteinases in the pathogenesis of a murine model of colitis. Am. J. Pathol. 157, 1927–1935 (2000).
Denson, L. A. et al. Clinical and genomic correlates of neutrophil reactive oxygen species production in pediatric patients with crohn’s disease. Gastroenterology 154, 2097–2110 (2018).
Biasi, F., Leonarduzzi, G., Oteiza, P. I. & Poli, G. Inflammatory bowel disease: mechanisms, redox considerations, and therapeutic targets. Antioxid. Redox Signal. 19, 1711–1747 (2013).
Karmakar, M. et al. N-GSDMD trafficking to neutrophil organelles facilitates IL-1β release independently of plasma membrane pores and pyroptosis. Nat. Commun. 11, 2212 (2020).
Tecchio, C. & Cassatella, M. A. Neutrophil-derived chemokines on the road to immunity. Semin. Immunol. 28, 119–128 (2016).
Pelletier, M. et al. Evidence for a cross-talk between human neutrophils and Th17 cells. Blood 115, 335–343 (2010).
Yang, D., Chen, Q., Chertov, O. & Oppenheim, J. J. Human neutrophil defensins selectively chemoattract naive T and immature dendritic cells. J. Leukoc. Biol. 68, 9–14 (2000).
Wang, S. et al. S100A8/A9 in inflammation. Front. Immunol. 9, 1298 (2018).
Li, T. et al. Neutrophil extracellular traps induce intestinal damage and thrombotic tendency in inflammatory bowel disease. J. Crohns Colitis 14, 240–253 (2020).
Leppkes, M. et al. Neutrophils prevent rectal bleeding in ulcerative colitis by peptidyl-arginine deiminase-4-dependent immunothrombosis. Gut 71, 2414–2429 (2022).
Kühl, A. A. et al. Aggravation of different types of experimental colitis by depletion or adhesion blockade of neutrophils. Gastroenterology 133, 1882–1892 (2007).
Zhang, R. et al. Up-regulation of Gr1+CD11b+ population in spleen of dextran sulfate sodium administered mice works to repair colitis. Inflamm. Allergy Drug. Targets 10, 39–46 (2011).
Nemoto, Y. et al. Negative feedback regulation of colitogenic CD4+ T cells by increased granulopoiesis. Inflamm. Bowel Dis. 14, 1491–1503 (2008).
Phillipson, M. & Kubes, P. The healing power of neutrophils. Trends Immunol. 40, 635–647 (2019).
Seo, D. H. et al. Triggering receptor expressed on myeloid cells-1 agonist regulates intestinal inflammation via Cd177+ neutrophils. Front. Immunol. 12, 650864 (2021).
Schwab, J. M., Chiang, N., Arita, M. & Serhan, C. N. Resolvin E1 and protectin D1 activate inflammation-resolution programmes. Nature 447, 869–874 (2007).
Ardi, V. C., Kupriyanova, T. A., Deryugina, E. I. & Quigley, J. P. Human neutrophils uniquely release TIMP-free MMP-9 to provide a potent catalytic stimulator of angiogenesis. Proc. Natl Acad. Sci. USA 104, 20262–20267 (2007).
Christoffersson, G. et al. VEGF-A recruits a proangiogenic MMP-9–delivering neutrophil subset that induces angiogenesis in transplanted hypoxic tissue. Blood 120, 4653–4662 (2012).
Lin, J. et al. Monocyte chemotactic protein 1-induced protein 1 is highly expressed in inflammatory bowel disease and negatively regulates neutrophil activities. Mediators Inflamm. 2020, 8812020 (2020).
Kienle, K. et al. Neutrophils self-limit swarming to contain bacterial growth in vivo. Science 372, eabe7729 (2021).
Bressenot, A. et al. Comparing histological activity indexes in UC. Gut 64, 1412–1418 (2015).
Demir, A. K. et al. The relationship between the neutrophil–lymphocyte ratio and disease activity in patients with ulcerative colitis. Kaohsiung J. Med. Sci. 31, 585–590 (2015).
Langley, B. O. et al. Inflammatory bowel disease and neutrophil–lymphocyte ratio: a systematic scoping review. J. Clin. Med. 10, 4219 (2021).
Dinallo, V. et al. Neutrophil extracellular traps sustain inflammatory signals in ulcerative colitis. J. Crohns Colitis 13, 772–784 (2019).
Herrero-Cervera, A., Soehnlein, O. & Kenne, E. Neutrophils in chronic inflammatory diseases. Cell. Mol. Immunol. 19, 177–191 (2022).
Sedghi, S. et al. Increased production of luminol enhanced chemiluminescence by the inflamed colonic mucosa in patients with ulcerative colitis. Gut 34, 1191–1197 (1993).
Zhang, T. et al. MK2 is required for neutrophil-derived ROS production and inflammatory bowel disease. Front. Med. 7, 207 (2020).
Liu, C. et al. Twist1 contributes to developing and sustaining corticosteroid resistance in ulcerative colitis. Theranostics 11, 7797–7812 (2021).
Lu, H. et al. Cyclosporine modulates neutrophil functions via the SIRT6–HIF‐1α–glycolysis axis to alleviate severe ulcerative colitis. Clin. Transl. Med. 11, e334 (2021).
Korzenik, J. R. & Dieckgraefe, B. K. Is Crohn’s disease due to defective immunity? Dig. Dis. Sci. 45, 1121–1129 (2000).
Hayee, B. et al. The neutrophil respiratory burst and bacterial digestion in Crohn’s disease. Dig. Dis. Sci. 56, 1482–1488 (2011).
Smith, A. M. et al. Disruption of macrophage pro-inflammatory cytokine release in Crohn’s disease is associated with reduced optineurin expression in a subset of patients. Immunology 144, 45–55 (2015).
Segal, A. W. The role of neutrophils in the pathogenesis of Crohn’s disease. Eur. J. Clin. Invest. 48, e12983 (2018).
Marks, D. J. B., Rahman, F. Z., Sewell, G. W. & Segal, A. W. Crohn’s disease: an immune deficiency state. Clin. Rev. Allergy Immunol. 38, 20–31 (2010).
Yu, H.-H., Yang, Y.-H. & Chiang, B.-L. Chronic granulomatous disease: a comprehensive review. Clin. Rev. Allergy Immunol. 61, 101–113 (2021).
Visser, G. et al. Neutropenia, neutrophil dysfunction, and inflammatory bowel disease in glycogen storage disease type Ib: results of the European Study on Glycogen Storage Disease Type I. J. Pediatr. 137, 187–191 (2000).
Britton, G. J. et al. Microbiotas from humans with inflammatory bowel disease alter the balance of gut Th17 and RORγt+ regulatory T cells and exacerbate colitis in mice. Immunity 50, 212–224.e4 (2019).
Lamas, B. et al. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat. Med. 22, 598–605 (2016).
Benech, N. & Sokol, H. Fecal microbiota transplantation in gastrointestinal disorders: time for precision medicine. Genome Med. 12, 58 (2020).
Danne, C., Rolhion, N. & Sokol, H. Recipient factors in faecal microbiota transplantation: one stool does not fit all. Nat. Rev. Gastroenterol. Hepatol. 18, 503–513 (2021).
Sokol, H. et al. Fecal microbiota transplantation to maintain remission in Crohn’s disease: a pilot randomized controlled study. Microbiome 8, 12 (2020).
D’Haens, G. R. & Jobin, C. Fecal microbial transplantation for diseases beyond recurrent Clostridium difficile infection. Gastroenterology 157, 624–636 (2019).
Darfeuille-Michaud, A. et al. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn’s disease. Gastroenterology 127, 412–421 (2004).
Knights, D. et al. Complex host genetics influence the microbiome in inflammatory bowel disease. Genome Med. 6, 107 (2014).
Sokol, H. et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl Acad. Sci. USA 105, 16731–16736 (2008).
Morgan, X. C. et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 13, R79 (2012).
Lloyd-Price, J. et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 569, 655–662 (2019).
Sokol, H. et al. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm. Bowel Dis. 15, 1183–1189 (2009).
Atarashi, K. et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 500, 232–236 (2013).
Arpaia, N. et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 504, 451–455 (2013).
Martín, R. et al. Faecalibacterium prausnitzii prevents physiological damages in a chronic low-grade inflammation murine model. BMC Microbiol. 15, 67 (2015).
Miquel, S. et al. Identification of metabolic signatures linked to anti-inflammatory effects of Faecalibacterium prausnitzii. mBio 6, e00300–e00315 (2015).
Quévrain, E. et al. Identification of an anti-inflammatory protein from Faecalibacterium prausnitzii, a commensal bacterium deficient in Crohn’s disease. Gut 65, 415–425 (2016).
Touch, S. et al. Human CD4+CD8α+ Tregs induced by Faecalibacterium prausnitzii protect against intestinal inflammation. JCI Insight 7, e154722 (2022).
Liu, J. Z. et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat. Genet. 47, 979–986 (2015).
GEM Project Research Consortium. Association of host genome with intestinal microbial composition in a large healthy cohort. Nat. Genet. 48, 1413–1417 (2016).
Bonder, M. J. et al. The effect of host genetics on the gut microbiome. Nat. Genet. 48, 1407–1412 (2016).
Wang, J. et al. Genome-wide association analysis identifies variation in vitamin D receptor and other host factors influencing the gut microbiota. Nat. Genet. 48, 1396–1406 (2016).
Huang, H. et al. Fine-mapping inflammatory bowel disease loci to single-variant resolution. Nature 547, 173–178 (2017).
de Lange, K. M. et al. Genome-wide association study implicates immune activation of multiple integrin genes in inflammatory bowel disease. Nat. Genet. 49, 256–261 (2017).
Aschard, H. et al. Genetic effects on the commensal microbiota in inflammatory bowel disease patients. PLOS Genet. 15, e1008018 (2019). This study highlights how genetics influence intestinal microbiota in IBD.
Chu, H. et al. Gene–microbiota interactions contribute to the pathogenesis of inflammatory bowel disease. Science 352, 1116–1120 (2016).
Lavelle, A. & Sokol, H. Gut microbiota-derived metabolites as key actors in inflammatory bowel disease. Nat. Rev. Gastroenterol. Hepatol. 17, 223–237 (2020). This review helps us to understand the role of microbial metabolites in IBD.
Roberts, R. L. et al. Confirmation of association of IRGM and NCF4 with ileal Crohn’s disease in a population-based cohort. Genes Immun. 9, 561–565 (2008).
Nuij, V. J. A. A., Peppelenbosch, M. P., van der Woude, C. J. & Fuhler, G. M. Genetic polymorphism in ATG16L1 gene is associated with adalimumab use in inflammatory bowel disease. J. Transl. Med. 15, 248 (2017).
Wright, M. et al. Early onset granulomatous colitis associated with a mutation in NCF4 resolved with hematopoietic stem cell transplantation. J. Pediatr. 210, 220–225 (2019).
Glennon-Alty, L., Moots, R. J., Edwards, S. W. & Wright, H. L. Type I interferon regulates cytokine-delayed neutrophil apoptosis, reactive oxygen species production and chemokine expression. Clin. Exp. Immunol. 203, 151–159 (2021).
Liu, Z. et al. Genetic architecture of the inflammatory bowel diseases across East Asian and European ancestries. Nat. Genet. 55, 796–806 (2023). Recent GWAS study in IBD that integrates individuals from non-European ancestries (specifically, individuals from East Asian ancestries).
Zhang, H., Meng, F., Chu, C.-L., Takai, T. & Lowell, C. A. The Src family kinases Hck and Fgr negatively regulate neutrophil and dendritic cell chemokine signaling via PIR-B. Immunity 22, 235–246 (2005).
Barrett, J. C. et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat. Genet. 40, 955–962 (2008).
Anderson, C. A. et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat. Genet. 43, 246–252 (2011).
Jostins, L. et al. Host–microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491, 119–124 (2012).
Liu, Z. et al. The kinase LRRK2 is a regulator of the transcription factor NFAT that modulates the severity of inflammatory bowel disease. Nat. Immunol. 12, 1063–1070 (2011).
Ahmadi Rastegar, D. & Dzamko, N. Leucine rich repeat kinase 2 and innate immunity. Front. Neurosci. 14, 193 (2020).
Panopoulos, A. D. et al. STAT3 governs distinct pathways in emergency granulopoiesis and mature neutrophils. Blood 108, 3682–3690 (2006).
Sazonovs, A. et al. Large-scale sequencing identifies multiple genes and rare variants associated with Crohn’s disease susceptibility. Nat. Genet. 54, 1275–1283 (2022). A recent study that analysed sequence data from more than 30,000 patients with Crohn’s disease and 80,000 population controls and identified new Crohn’s disease susceptibility genes.
Turpin, W., Goethel, A., Bedrani, L. & Croitoru, Mdcm,K. Determinants of IBD heritability: genes, bugs, and more. Inflamm. Bowel Dis. 24, 1133–1148 (2018).
Russo, I., Bubacco, L. & Greggio, E. LRRK2 as a target for modulating immune system responses. Neurobiol. Dis. 169, 105724 (2022).
Le, N. P. K., Channabasappa, S., Hossain, M., Liu, L. & Singh, B. Leukocyte-specific protein 1 regulates neutrophil recruitment in acute lung inflammation. Am. J. Physiol. Lung Cell. Mol. Physiol. 309, L995–L1008 (2015).
O’Sullivan, S., Gilmer, J. F. & Medina, C. Matrix metalloproteinases in inflammatory bowel disease: an update. Mediators Inflamm. 2015, 964131 (2015).
Wang, Y. & Jönsson, F. Expression, role, and regulation of neutrophil Fcγ receptors. Front. Immunol. 10, 1958 (2019).
Miyauchi, M. et al. Efficient production of human neutrophils from iPSCs that prevent murine lethal infection with immune cell recruitment. Blood 138, 2555–2569 (2021).
Danne, C. et al. CARD9 in neutrophils protects from colitis and controls mitochondrial metabolism and cell survival. Gut 72, 1081–1092 (2022). A study that demonstrates new roles for CARD9, an IBD susceptibility gene, in neutrophil functionality.
Mirkov, M. U., Verstockt, B. & Cleynen, I. Genetics of inflammatory bowel disease: beyond NOD2. Lancet Gastroenterol. Hepatol. 2, 224–234 (2017).
Sokol, H. et al. Intestinal dysbiosis in inflammatory bowel disease associated with primary immunodeficiency. J. Allergy Clin. Immunol. 143, 775–778.e6 (2019).
Castagnoli, R. et al. Gut microbiota–host interactions in inborn errors of immunity. Int. J. Mol. Sci. 22, 1416 (2021).
Sokol, H. et al. Fungal microbiota dysbiosis in IBD. Gut 66, 1039–1048 (2017).
Belkaid, Y. & Harrison, O. J. Homeostatic immunity and the microbiota. Immunity 46, 562–576 (2017).
Molloy, M. J. et al. Intraluminal containment of commensal outgrowth in the gut during infection-induced dysbiosis. Cell Host Microbe 14, 318–328 (2013).
Spehlmann, M. E. et al. CXCR2-dependent mucosal neutrophil influx protects against colitis-associated diarrhea caused by an attaching/effacing lesion-forming bacterial pathogen. J. Immunol. 183, 3332–3343 (2009).
Watanabe, K. et al. Microbiome-mediated neutrophil recruitment via CXCR2 and protection from amebic colitis. PLoS Pathog. 13, e1006513 (2017).
Flannigan, K. L. et al. IL-17A-mediated neutrophil recruitment limits expansion of segmented filamentous bacteria. Mucosal Immunol. 10, 673–684 (2017).
Waldschmitt, N. et al. The regenerating family member 3β instigates IL-17A-mediated neutrophil recruitment downstream of NOD1/2 signalling for controlling colonisation resistance independently of microbiota community structure. Gut 68, 1190–1199 (2019).
Jee, J. et al. Cxcr2 signaling and the microbiome suppress inflammation, bile duct injury, and the phenotype of experimental biliary atresia. PLoS ONE 12, e0182089 (2017).
Mei, J. et al. Cxcr2 and Cxcl5 regulate the IL-17/G-CSF axis and neutrophil homeostasis in mice. J. Clin. Invest. 122, 974–986 (2012).
Zindl, C. L. et al. IL-22–producing neutrophils contribute to antimicrobial defense and restitution of colonic epithelial integrity during colitis. Proc. Natl Acad. Sci. USA 110, 12768–12773 (2013).
Kamada, N. & Núñez, G. Role of the gut microbiota in the development and function of lymphoid cells. J. Immunol. 190, 1389–1395 (2013).
Winter, S. E. et al. Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science 339, 708–711 (2013).
Zechner, E. L. Inflammatory disease caused by intestinal pathobionts. Curr. Opin. Microbiol. 35, 64–69 (2017).
Winter, S. E. et al. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467, 426–429 (2010).
Gubatan, J. et al. Antimicrobial peptides and the gut microbiome in inflammatory bowel disease. World J. Gastroenterol. 27, 7402–7422 (2021).
Cullen, T. W. et al. Antimicrobial peptide resistance mediates resilience of prominent gut commensals during inflammation. Science 347, 170–175 (2015).
Yoshimura, T. et al. The antimicrobial peptide CRAMP is essential for colon homeostasis by maintaining microbiota balance. J. Immunol. 200, 2174–2185 (2018).
Gill, N. et al. Neutrophil elastase alters the murine gut microbiota resulting in enhanced salmonella colonization. PLoS ONE 7, e49646 (2012).
Raffatellu, M. et al. Lipocalin-2 resistance confers an advantage to Salmonella enterica serotype Typhimurium for growth and survival in the inflamed intestine. Cell Host Microbe 5, 476–486 (2009).
Liu, J. Z. et al. Zinc sequestration by the neutrophil protein calprotectin enhances Salmonella growth in the inflamed gut. Cell Host Microbe 11, 227–239 (2012).
Fischbach, M. A., Lin, H., Liu, D. R. & Walsh, C. T. How pathogenic bacteria evade mammalian sabotage in the battle for iron. Nat. Chem. Biol. 2, 132–138 (2006).
Deriu, E. et al. Probiotic bacteria reduce Salmonella Typhimurium intestinal colonization by competing for iron. Cell Host Microbe 14, 26–37 (2013).
Loh, J. T. et al. DOK3 maintains intestinal homeostasis by suppressing JAK2/STAT3 signaling and S100a8/9 production in neutrophils. Cell Death Dis. 12, 1054 (2021).
Ilott, N. E. et al. Defining the microbial transcriptional response to colitis through integrated host and microbiome profiling. ISME J. 10, 2389–2404 (2016).
Geva-Zatorsky, N. et al. Mining the human gut microbiota for immunomodulatory organisms. Cell 168, 928–943.e11 (2017).
Martini, E., Krug, S. M., Siegmund, B., Neurath, M. F. & Becker, C. Mend your fences. Cell. Mol. Gastroenterol. Hepatol. 4, 33–46 (2017).
Nishida, A. et al. Gut microbiota in the pathogenesis of inflammatory bowel disease. Clin. J. Gastroenterol. 11, 1–10 (2018).
Fang, X., Li, F. & Hong, D. Potential role of Akkermansia muciniphila in Parkinson’s disease and other neurological/autoimmune diseases. Curr. Med. Sci. 41, 1172–1177 (2021).
Goris, H., de Boer, F. & van der Waaij, D. Myelopoiesis in experimentally contaminated specific-pathogen-free and germfree mice during oral administration of polymyxin. Infect. Immun. 50, 437–441 (1985).
Tada, T., Yamamura, S., Kuwano, Y. & Abo, T. Level of myelopoiesis in the bone marrow is influenced by intestinal flora. Cell. Immunol. 173, 155–161 (1996).
Zhang, H., Sparks, J. B., Karyala, S. V., Settlage, R. & Luo, X. M. Host adaptive immunity alters gut microbiota. ISME J. 9, 770–781 (2015).
Khosravi, A. et al. Gut microbiota promote hematopoiesis to control bacterial infection. Cell Host Microbe 15, 374–381 (2014).
Deshmukh, H. S. et al. The microbiota regulates neutrophil homeostasis and host resistance to Escherichia coli K1 sepsis in neonatal mice. Nat. Med. 20, 524–530 (2014).
Josefsdottir, K. S., Baldridge, M. T., Kadmon, C. S. & King, K. Y. Antibiotics impair murine hematopoiesis by depleting the intestinal microbiota. Blood 129, 729–739 (2017).
Balmer, M. L. et al. Microbiota-derived compounds drive steady-state granulopoiesis via MyD88/TICAM signaling. J. Immunol. 193, 5273–5283 (2014).
Brenchley, J. M. & Douek, D. C. Microbial translocation across the GI tract. Annu. Rev. Immunol. 30, 149–173 (2012).
Clarke, T. B. et al. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat. Med. 16, 228–231 (2010).
Iwamura, C., Bouladoux, N., Belkaid, Y., Sher, A. & Jankovic, D. Sensing of the microbiota by NOD1 in mesenchymal stromal cells regulates murine hematopoiesis. Blood 129, 171–176 (2017).
Burgess, S. L. et al. Gut microbiome communication with bone marrow regulates susceptibility to amebiasis. J. Clin. Invest. 130, 4019–4024 (2020).
Murdoch, C. C. et al. Intestinal serum amyloid A suppresses systemic neutrophil activation and bactericidal activity in response to microbiota colonization. PLOS Pathog. 15, e1007381 (2019).
Ascher, S. et al. Gut microbiota restricts NETosis in acute mesenteric ischemia-reperfusion injury. Arterioscler. Thromb. Vasc. Biol. 40, 2279–2292 (2020).
Pieterse, E., Rother, N., Yanginlar, C., Hilbrands, L. B. & van der Vlag, J. Neutrophils discriminate between lipopolysaccharides of different bacterial sources and selectively release neutrophil extracellular traps. Front. Immunol. 7, 484 (2016).
Vong, L., Lorentz, R. J., Assa, A., Glogauer, M. & Sherman, P. M. Probiotic Lactobacillus rhamnosus inhibits the formation of neutrophil extracellular traps. J. Immunol. 192, 1870–1877 (2014).
Roselli, M., Finamore, A., Britti, M. S. & Mengheri, E. Probiotic bacteria Bifidobacterium animalis MB5 and Lactobacillus rhamnosus GG protect intestinal Caco-2 cells from the inflammation-associated response induced by enterotoxigenic Escherichia coli K88. Br. J. Nutr. 95, 1177–1184 (2006).
Martin, C. et al. Chemokines acting via CXCR2 and CXCR4 control the release of neutrophils from the bone marrow and their return following senescence. Immunity 19, 583–593 (2003).
Rankin, S. M. The bone marrow: a site of neutrophil clearance. J. Leukoc. Biol. 88, 241–251 (2010).
Schulthess, J. et al. The short chain fatty acid butyrate imprints an antimicrobial program in macrophages. Immunity 50, 432–445.e7 (2019).
Íñiguez-Gutiérrez, L. et al. Physiological concentrations of short-chain fatty acids induce the formation of neutrophil extracellular traps in vitro. Int. J. Immunopathol. Pharmacol. 34, 205873842095894 (2020).
Maslowski, K. M. et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature 461, 1282–1286 (2009). One of the first studies to show the effect of a SCFA on neutrophil function.
Aoyama, M., Kotani, J. & Usami, M. Butyrate and propionate induced activated or non-activated neutrophil apoptosis via HDAC inhibitor activity but without activating GPR-41/GPR-43 pathways. Nutrition 26, 653–661 (2010).
Li, G. et al. Microbiota metabolite butyrate constrains neutrophil functions and ameliorates mucosal inflammation in inflammatory bowel disease. Gut Microbes 13, 1968257 (2021). An interesting study that focuses on a microbial metabolite and neutrophils in IBD.
Vinolo, M. A. R. et al. Short-chain fatty acids stimulate the migration of neutrophils to inflammatory sites. Clin. Sci. 117, 331–338 (2009).
Vinolo, M. A. R., Hatanaka, E., Lambertucci, R. H., Newsholme, P. & Curi, R. Effects of short chain fatty acids on effector mechanisms of neutrophils. Cell Biochem. Funct. 27, 48–55 (2009).
Alexeev, E. E. et al. Microbial‐derived indoles inhibit neutrophil myeloperoxidase to diminish bystander tissue damage. FASEB J. 35, e21552 (2021). This study reveals an inhibiting effect of indoles on neutrophil MPO activity.
Santoro, P. et al. Unconjugated bile acids modulate adult and neonatal neutrophil chemotaxis induced in vitro by N-formyl-Met-Leu-Phe-peptide. Pediatr. Res. 51, 392–396 (2002).
Balazs, I. et al. Serum bile acids in liver cirrhosis promote neutrophil dysfunction. Clin. Transl. Med. 12, e735 (2022). This human study highlights the effect of microbiota-derived bile acids on neutrophil function.
Zelante, T. et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity 39, 372–385 (2013).
Ye, J. et al. The aryl hydrocarbon receptor preferentially marks and promotes gut regulatory T cells. Cell Rep. 21, 2277–2290 (2017).
Koeth, R. A. et al. Intestinal microbiota metabolism of l-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat. Med. 19, 576–585 (2013).
Levy, M. et al. Microbiota-modulated metabolites shape the intestinal microenvironment by regulating NLRP6 inflammasome signaling. Cell 163, 1428–1443 (2015).
Zhu, W. et al. Gut microbial metabolite TMAO enhances platelet hyperreactivity and thrombosis risk. Cell 165, 111–124 (2016).
José Fábrega, M. et al. Activation of immune and defense responses in the intestinal mucosa by outer membrane vesicles of commensal and probiotic escherichia coli strains. Front. Microbiol. 7, 705 (2016).
Lajqi, T. et al. Gut microbiota-derived small extracellular vesicles endorse memory-like inflammatory responses in murine neutrophils. Biomedicines 10, 442 (2022).
Brok-Volchanskaya, V. S. et al. Effective and rapid generation of functional neutrophils from induced pluripotent stem cells using ETV2-modified mRNA. Stem Cell Rep. 13, 1099–1110 (2019).
Németh, T., Sperandio, M. & Mócsai, A. Neutrophils as emerging therapeutic targets. Nat. Rev. Drug. Discov. 19, 253–275 (2020).
Krupa, A. et al. Silencing Bruton’s tyrosine kinase in alveolar neutrophils protects mice from LPS/immune complex-induced acute lung injury. Am. J. Physiol. Lung Cell. Mol. Physiol. 307, L435–L448 (2014).
Reshetnikov, V. et al. Chemical tools for targeted amplification of reactive oxygen species in neutrophils. Front. Immunol. 9, 1827 (2018).
Han, X. et al. Loss of GM-CSF signalling in non-haematopoietic cells increases NSAID ileal injury. Gut 59, 1066–1078 (2010).
Däbritz, J. Granulocyte macrophage colony-stimulating factor and the intestinal innate immune cell homeostasis in Crohn’s disease. Am. J. Physiol. Gastrointest. Liver Physiol. 306, G455–G465 (2014).
Dieckgraefe, B. K. & Korzenik, J. R. Treatment of active Crohn’s disease with recombinant human granulocyte-macrophage colony-stimulating factor. Lancet 360, 1478–1480 (2002).
Korzenik, J. R., Dieckgraefe, B. K., Valentine, J. F., Hausman, D. F. & Gilbert, M. J. Sargramostim for active Crohn’s disease. N. Engl. J. Med. 352, 2193–2201 (2005).
Roth, L., MacDonald, J. K., McDonald, J. W. D. & Chande, N. Sargramostim (GM-CSF) for Induction of remission in Crohnʼs disease: a cochrane inflammatory bowel disease and functional bowel disorders systematic review of randomized trials. Inflamm. Bowel Dis. 18, 1333–1339 (2012).
Hueber, W. et al. Secukinumab, a human anti-IL-17A monoclonal antibody, for moderate to severe Crohn’s disease: unexpected results of a randomised, double-blind placebo-controlled trial. Gut 61, 1693–1700 (2012).
Sandborn, W. J. et al. Andecaliximab [anti-matrix metalloproteinase-9] induction therapy for ulcerative colitis: a randomised, double-blind, placebo-controlled, phase 2/3 study in patients with moderate to severe disease. J. Crohns Colitis 12, 1021–1029 (2018).
Schreiber, S. et al. A phase 2, randomized, placebo-controlled study evaluating matrix metalloproteinase-9 inhibitor, andecaliximab, in patients with moderately to severely active crohn’s disease. J. Crohns Colitis 12, 1014–1020 (2018).
Rahman et al. Phagocyte dysfunction and inflammatory bowel disease. Inflamm. Bowel Dis. 14, 1443–1452 (2008).
Author information
Authors and Affiliations
Contributions
All authors contributed equally to all aspects of the manuscript.
Corresponding author
Ethics declarations
Competing interests
H.S. reports lecture fees, board membership or consultancy from Amgen, Fresenius, IPSEN, Actial, Astellas, Danone, THAC, Biose, BiomX, Eligo, Immusmol, Adare, Nestlé, Ferring, MSD, Bledina, Pfizer, Biocodex, BMS, Bromatech, Gilead, Janssen, Mayoli, Roche, Sanofi, Servier, Takeda, AbbVie, has stocks from Enterome Bioscience and is co-founder of Exeliom Biosciences. The other authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Gastroenterology & Hepatology thanks Zhanju Liu, Britta Siegmund and Andrew Gewirtz for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Ensembl version 103.38: http://www.ensembl.org/index.html
Human Protein Atlas version 22.0: https://www.proteinatlas.org/
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Danne, C., Skerniskyte, J., Marteyn, B. et al. Neutrophils: from IBD to the gut microbiota. Nat Rev Gastroenterol Hepatol 21, 184–197 (2024). https://doi.org/10.1038/s41575-023-00871-3
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41575-023-00871-3