Abstract
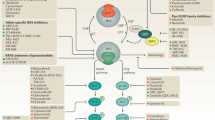
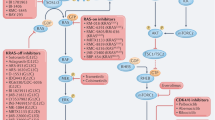
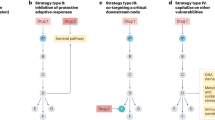
RAS genes are the most commonly mutated oncogenes in cancer, but effective therapeutic strategies to target RAS-mutant cancers have proved elusive. A key aspect of this challenge is the fact that direct inhibition of RAS proteins has proved difficult, leading researchers to test numerous alternative strategies aimed at exploiting RAS-related vulnerabilities or targeting RAS effectors. In the past few years, we have witnessed renewed efforts to target RAS directly, with several promising strategies being tested in clinical trials at different stages of completion. Important advances have also been made in approaches designed to indirectly target RAS by improving inhibition of RAS effectors, exploiting synthetic lethal interactions or metabolic dependencies, using therapeutic combination strategies or harnessing the immune system. In this Review, we describe historical and ongoing efforts to target RAS-mutant cancers and outline the current therapeutic landscape in the collective quest to overcome the effects of this crucial oncogene.
Key points
-
RAS is the most commonly mutated oncogene in cancer.
-
Direct inhibition of oncogenic RAS has proved difficult, hindering the development of effective therapies for RAS-mutant cancers.
-
Renewed efforts to target mutant RAS directly have yielded promising efficacy outcomes in preclinical studies.
-
Targeting key effector pathways or vulnerabilities in RAS-mutant cancers offers a promising alternative therapeutic approach.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Simanshu, D. K., Nissley, D. V. & McCormick, F. RAS proteins and their regulators in human disease. Cell 170, 17–33 (2017).
Vetter, I. R. & Wittinghofer, A. The guanine nucleotide-binding switch in three dimensions. Science 294, 1299–1304 (2001).
Nussinov, R., Tsai, C.-J. & Jang, H. Oncogenic Ras isoforms signaling specificity at the membrane. Cancer Res. 78, 593–602 (2018).
Keeton, A. B., Salter, E. A. & Piazza, G. A. The RAS-effector interaction as a drug target. Cancer Res. 77, 221–226 (2017).
Ryan, M. B., Der, C. J., Wang-Gillam, A. & Cox, A. D. Targeting RAS-mutant cancers: is ERK the key? Trends Cancer 1, 183–198 (2015).
Fruman, D. A. et al. The PI3K pathway in human disease. Cell 170, 605–635 (2017).
Chow, H. Y. et al. p21-activated kinase 1 is required for efficient tumor formation and progression in a Ras-mediated skin cancer model. Cancer Res. 72, 5966–5975 (2012).
Kashatus, D. F. Ral GTPases in tumorigenesis: emerging from the shadows. Exp. Cell Res. 319, 2337–2342 (2013).
Zhu, Z. et al. Inhibition of KRAS-driven tumorigenicity by interruption of an autocrine cytokine circuit. Cancer Discov. 4, 452–465 (2014).
Yuan, T. L. et al. Differential effector engagement by oncogenic KRAS. Cell Rep. 22, 1889–1902 (2018).
Cox, A. D. & Der, C. J. Ras history: the saga continues. Small GTPases 1, 2–27 (2010).
Vogelstein, B. et al. Cancer genome landscapes. Science 339, 1546–1558 (2013).
Forbes, S. A. et al. COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res. 45, D777–D783 (2017).
Wellcome Sanger Institute. COSMIC Release v84. Sanger.ac.uk https://cosmic-blog.sanger.ac.uk/cosmic-release-v84/ (2018).
Hobbs, G. A., Der, C. J. & Rossman, K. L. RAS isoforms and mutations in cancer at a glance. J. Cell Sci. 129, 1287–1292 (2016).
Burd, C. E. et al. Mutation-specific RAS oncogenicity explains NRAS codon 61 selection in melanoma. Cancer Discov. 4, 1418–1429 (2014).
Haigis, K. M. et al. Differential effects of oncogenic K-Ras and N-Ras on proliferation, differentiation and tumor progression in the colon. Nat. Genet. 40, 600–608 (2008).
Raphael, B. J. et al. Integrated genomic characterization of pancreatic ductal adenocarcinoma. Cancer Cell 32, 185–203.e13 (2017).
Collisson, E. A. et al. A central role for RAF-MEK-ERK signaling in the genesis of pancreatic ductal adenocarcinoma. Cancer Discov. 2, 685–693 (2012).
Appleman, V. A., Ahronian, L. G., Cai, J., Klimstra, D. S. & Lewis, B. C. KRAS(G12D)- and BRAF(V600E)-induced transformation of murine pancreatic epithelial cells requires MEK/ERK-stimulated IGF1R signaling. Mol. Cancer Res. 10, 1228–1239 (2012).
Collins, M. A. et al. Oncogenic Kras is required for both the initiation and maintenance of pancreatic cancer in mice. J. Clin. Invest. 122, 639–653 (2012).
Esposito, D., Stephen, A. G., Turbyville, T. J. & Holderfield, M. New weapons to penetrate the armor: novel reagents and assays developed at the NCI RAS Initiative to enable discovery of RAS therapeutics. Semin. Cancer Biol. https://doi.org/10.1016/j.semcancer.2018.02.006 (2018).
Waters, A. M. et al. Evaluation of the selectivity and sensitivity of isoform- and mutation-specific RAS antibodies. Sci. Signal. 10, eaao3332 (2017).
Goody, R. S., Frech, M. & Wittinghofer, A. Affinity of guanine nucleotide binding proteins for their ligands: facts and artefacts. Trends Biochem. Sci. 16, 327–328 (1991).
Ahearn, I. M., Haigis, K., Bar-Sagi, D. & Philips, M. R. Regulating the regulator: post-translational modification of RAS. Nat. Rev. Mol. Cell Biol. 13, 39–51 (2011).
Basso, A. D., Kirschmeier, P. & Bishop, W. R. Lipid posttranslational modifications. Farnesyl transferase inhibitors. J. Lipid Res. 47, 15–31 (2006).
Berndt, N., Hamilton, A. D. & Sebti, S. M. Targeting protein prenylation for cancer therapy. Nat. Rev. Cancer 11, 775–791 (2011).
Rowell, C. A., Kowalczyk, J. J., Lewis, M. D. & Garcia, A. M. Direct demonstration of geranylgeranylation and farnesylation of Ki-Ras in vivo. J. Biol. Chem. 272, 14093–14097 (1997).
Whyte, D. B. et al. K− and N-Ras are geranylgeranylated in cells treated with farnesyl protein transferase inhibitors. J. Biol. Chem. 272, 14459–14464 (1997).
Liu, M. et al. Targeting the protein prenyltransferases efficiently reduces tumor development in mice with K-RAS-induced lung cancer. Proc. Natl Acad. Sci. USA 107, 6471–6476 (2010).
Winter-Vann, A. M. & Casey, P. J. Post-prenylation-processing enzymes as new targets in oncogenesis. Nat. Rev. Cancer 5, 405–412 (2005).
Zimmermann, G. et al. Small molecule inhibition of the KRAS-PDEdelta interaction impairs oncogenic KRAS signalling. Nature 497, 638–642 (2013).
Papke, B. et al. Identification of pyrazolopyridazinones as PDEdelta inhibitors. Nat. Commun. 7, 11360 (2016).
Wang, M. & Casey, P. J. Protein prenylation: unique fats make their mark on biology. Nat. Rev. Mol. Cell Biol. 17, 110 (2016).
Cox, A. D., Fesik, S. W., Kimmelman, A. C., Luo, J. & Der, C. J. Drugging the undruggable RAS: mission possible? Nat. Rev. Drug Discov. 13, 828–851 (2014).
Hunter, J. C. et al. Biochemical and structural analysis of common cancer-associated KRAS mutations. Mol. Cancer Res. 13, 1325–1335 (2015).
Ostrem, J. M., Peters, U., Sos, M. L., Wells, J. A. & Shokat, K. M. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature 503, 548–551 (2013).
Hunter, J. C. et al. In situ selectivity profiling and crystal structure of SML-8-73-1, an active site inhibitor of oncogenic K-Ras G12C. Proc. Natl Acad. Sci. USA 111, 8895–8900 (2014).
Visscher, M., Arkin, M. R. & Dansen, T. B. Covalent targeting of acquired cysteines in cancer. Curr. Opin. Chem. Biol. 30, 61–67 (2016).
Shipman, L. Anticancer drugs: putting the brakes on KRAS-G12C nucleotide cycling. Nat. Rev. Drug Discov. 15, 159 (2016).
Westover, K. D., Janne, P. A. & Gray, N. S. Progress on covalent inhibition of KRAS(G12C). Cancer Discov. 6, 233–234 (2016).
Lito, P., Solomon, M., Li, L. S., Hansen, R. & Rosen, N. Allele-specific inhibitors inactivate mutant KRAS G12C by a trapping mechanism. Science 351, 604–608 (2016).
Patricelli, M. P. et al. Selective inhibition of oncogenic KRAS output with small molecules targeting the inactive state. Cancer Discov. 6, 316–329 (2016).
Ostrem, J. M. & Shokat, K. M. Direct small-molecule inhibitors of KRAS: from structural insights to mechanism-based design. Nat. Rev. Drug Discov. 15, 771–785 (2016).
Zeng, M. et al. Potent and selective covalent quinazoline inhibitors of KRAS G12C. Cell Chem. Biol. 24, 1005–1016 (2017).
Janes, M. R. et al. Targeting KRAS mutant cancers with a covalent G12C-specific inhibitor. Cell 172, 578–589.e17 (2018).
Welsch, M. E. et al. Multivalent small-molecule pan-RAS inhibitors. Cell 168, 878–889 (2017).
Kamerkar, S. et al. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature 546, 498–503 (2017).
Ross, S. J. et al. Targeting KRAS-dependent tumors with AZD4785, a high-affinity therapeutic antisense oligonucleotide inhibitor of KRAS. Sci. Transl Med. 9, eaal5253 (2017).
Athuluri-Divakar, S. K. et al. A small molecule RAS-mimetic disrupts RAS association with effector proteins to block signaling. Cell 165, 643–655 (2016).
Ritt, D. A. et al. Inhibition of Ras/Raf/MEK/ERK pathway signaling by a stress-induced phospho-regulatory circuit. Mol. Cell 64, 875–887 (2016).
Jost, M. et al. Combined CRISPRi/a-based chemical genetic screens reveal that rigosertib is a microtubule-destabilizing agent. Mol. Cell 68, 210–223 (2017).
Burns, M. C. et al. Approach for targeting Ras with small molecules that activate SOS-mediated nucleotide exchange. Proc. Natl Acad. Sci. USA 111, 3401–3406 (2014).
Burns, M. C. et al. High-throughput screening identifies small molecules that bind to the RAS:SOS:RAS complex and perturb RAS signaling. Anal. Biochem. 548, 44–52 (2018).
Gentile, D. R. et al. Ras binder induces a modified switch-II pocket in GTP and GDP states. Cell Chem. Biol. 24, 1455–1466.e14 (2017).
Lin, W.-C. et al. H-Ras forms dimers on membrane surfaces via a protein–protein interface. Proc. Natl Acad. Sci. 111, 2996–3001 (2014).
Santos, E. Dimerization opens new avenues into Ras signaling research. Sci. Signal. 7, pe12 (2014).
Nan, X. et al. Ras-GTP dimers activate the mitogen-activated protein kinase (MAPK) pathway. Proc. Natl Acad. Sci. USA 112, 7996–8001 (2015).
Sarkar-Banerjee, S. et al. Spatiotemporal analysis of K-Ras plasma membrane interactions reveals multiple high order homo-oligomeric complexes. J. Am. Chem. Soc. 139, 13466–13475 (2017).
Ambrogio, C. et al. KRAS dimerization impacts MEK inhibitor sensitivity and oncogenic activity of mutant KRAS. Cell 172, 857–868 (2018).
Spencer-Smith, R. et al. Inhibition of RAS function through targeting an allosteric regulatory site. Nat. Chem. Biol. 13, 62–68 (2017).
Freeman, A. K., Ritt, D. A. & Morrison, D. K. The importance of Raf dimerization in cell signaling. Small GTPases 4, 180–185 (2013).
Peng, S. B. et al. Inhibition of RAF isoforms and active dimers by LY3009120 leads to anti-tumor activities in RAS or BRAF mutant cancers. Cancer Cell 28, 384–398 (2015).
Lavoie, H. & Therrien, M. Regulation of RAF protein kinases in ERK signalling. Nat. Rev. Mol. Cell Biol. 16, 281–298 (2015).
Young, A., Lou, D. & McCormick, F. Oncogenic and wild-type Ras play divergent roles in the regulation of mitogen-activated protein kinase signaling. Cancer Discov. 3, 112–123 (2013).
Grabocka, E. et al. Wild-Type H− and N-Ras promote mutant K-Ras-driven tumorigenesis by modulating the DNA damage response. Cancer Cell 25, 243–256 (2014).
Singh, A. et al. A gene expression signature associated with “K-Ras addiction” reveals regulators of EMT and tumor cell survival. Cancer Cell 15, 489–500 (2009).
Muzumdar, M. D. et al. Survival of pancreatic cancer cells lacking KRAS function. Nat. Commun. 8, 1090 (2017).
Downward, J. RAS synthetic lethal screens revisited: still seeking the elusive prize? Clin. Cancer Res. 21, 1802–1809 (2015).
Scholl, C. et al. Synthetic lethal interaction between oncogenic KRAS DEPENDENcy and STK33 suppression in human cancer cells. Cell 137, 821–834 (2009).
Barbie, D. A. et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 462, 108–112 (2009).
Babij, C. et al. STK33 kinase activity is nonessential in KRAS-dependent cancer cells. Cancer Res. 71, 5818–5826 (2011).
Luo, T. et al. STK33 kinase inhibitor BRD-8899 has no effect on KRAS-dependent cancer cell viability. Proc. Natl Acad. Sci. USA 109, 2860–2865 (2012).
Muvaffak, A. et al. Evaluating TBK1 as a therapeutic target in cancers with activated IRF3. Mol. Cancer Res. 12, 1055–1066 (2014).
Wang, T. et al. Gene essentiality profiling reveals gene networks and synthetic lethal interactions with oncogenic Ras. Cell 168, 890–903.e15 (2017).
Anderson, G. R. et al. A landscape of therapeutic cooperativity in KRAS mutant cancers reveals principles for controlling tumor evolution. Cell Rep. 20, 999–1015 (2017).
Martin, T. D. et al. A role for mitochondrial translation in promotion of viability in K-Ras mutant cells. Cell Rep. 20, 427–438 (2017).
Martz, C. A. et al. Systematic identification of signaling pathways with potential to confer anticancer drug resistance. Sci. Signal. 7, ra121 (2014).
Kimmelman, A. C. Metabolic dependencies in RAS-driven cancers. Clin. Cancer Res. 21, 1828–1834 (2015).
Bryant, K. L., Mancias, J. D., Kimmelman, A. C. & Der, C. J. KRAS: feeding pancreatic cancer proliferation. Trends Biochem. Sci. 39, 91–100 (2014).
Commisso, C. et al. Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature 497, 633–637 (2013).
Son, J. et al. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature 496, 101–105 (2013).
Yang, S. et al. Pancreatic cancers require autophagy for tumor growth. Genes Dev. 25, 717–729 (2011).
Ying, H. et al. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell 149, 656–670 (2012).
Gaglio, D. et al. Oncogenic K-Ras decouples glucose and glutamine metabolism to support cancer cell growth. Mol. Syst. Biol. 7, 523–523 (2011).
Halbrook, C. J. & Lyssiotis, C. A. Employing metabolism to improve the diagnosis and treatment of pancreatic cancer. Cancer Cell 31, 5–19 (2017).
Gross, M. I. et al. Antitumor activity of the glutaminase inhibitor CB-839 in triple-negative breast cancer. Mol. Cancer Ther. 13, 890–901 (2014).
Jacque, N. et al. Targeting glutaminolysis has antileukemic activity in acute myeloid leukemia and synergizes with BCL-2 inhibition. Blood 126, 1346–1356 (2015).
Chakrabarti, G. et al. Targeting glutamine metabolism sensitizes pancreatic cancer to PARP-driven metabolic catastrophe induced by ß-lapachone. Cancer Metab. 3, 12 (2015).
Yun, J. et al. Vitamin C selectively kills KRAS and BRAF mutant colorectal cancer cells by targeting GAPDH. Science 350, 1391–1396 (2015).
Morrison, D. K. MAP kinase pathways. Cold Spring Harb. Perspect Biol. 4, a011254 (2012).
Blasco, R. B. et al. c-Raf, but not B-Raf, is essential for development of K-Ras oncogene-driven non-small cell lung carcinoma. Cancer Cell 19, 652–663 (2011).
Sanclemente, M. et al. c-RAF ablation induces regression of advanced Kras/Trp53 mutant lung adenocarcinomas by a mechanism independent of MAPK signaling. Cancer Cell 33, 217–228 (2018).
Nazarian, R. et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature 468, 973–977 (2010).
Johannessen, C. M. et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature 468, 968–972 (2010).
Ahronian, L. G. et al. Clinical acquired resistance to RAF inhibitor combinations in BRAF-mutant colorectal cancer through MAPK pathway alterations. Cancer Discov. 5, 358–367 (2015).
Oddo, D. et al. Molecular landscape of acquired resistance to targeted therapy combinations in BRAF-mutant colorectal cancer. Cancer Res. 76, 4504–4515 (2016).
Lidsky, M. et al. Mitogen-activated protein kinase (MAPK) hyperactivation and enhanced NRAS expression drive acquired vemurafenib resistance in V600E BRAF melanoma cells. J. Biol. Chem. 289, 27714–27726 (2014).
Wagle, N. et al. MAP kinase pathway alterations in BRAF-mutant melanoma patients with acquired resistance to combined RAF/MEK inhibition. Cancer Discov. 4, 61–68 (2014).
Van Allen, E. M. et al. The genetic landscape of clinical resistance to RAF inhibition in metastatic melanoma. Cancer Discov. 4, 94–109 (2014).
Watson, I. R. et al. The RAC1 P29S hotspot mutation in melanoma confers resistance to pharmacological inhibition of RAF. Cancer Res. 74, 4845–4852 (2014).
Corcoran, R. B. et al. BRAF gene amplification can promote acquired resistance to MEK inhibitors in cancer cells harboring the BRAF V600E mutation. Sci. Signal. 3, ra84 (2010).
Poulikakos, P. I., Zhang, C., Bollag, G., Shokat, K. M. & Rosen, N. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature 464, 427–430 (2010).
Callahan, M. K. et al. Progression of RAS-mutant leukemia during RAF inhibitor treatment. N. Engl. J. Med. 367, 2316–2321 (2012).
Oberholzer, P. A. et al. RAS mutations are associated with the development of cutaneous squamous cell tumors in patients treated with RAF inhibitors. J. Clin. Oncol. 30, 316–321 (2012).
Hatzivassiliou, G. et al. Mechanism of MEK inhibition determines efficacy in mutant KRAS- versus BRAF-driven cancers. Nature 501, 232–236 (2013).
Lito, P. et al. Disruption of CRAF-mediated MEK activation is required for effective MEK inhibition in KRAS mutant tumors. Cancer Cell 25, 697–710 (2014).
Wee, S. et al. PI3K pathway activation mediates resistance to MEK inhibitors in KRAS mutant cancers. Cancer Res. 69, 4286–4293 (2009).
Poulikakos, P. I. & Solit, D. B. Resistance to MEK inhibitors: should we co-target upstream? Sci. Signal. 4, pe16 (2011).
Sun, C. et al. Intrinsic resistance to MEK inhibition in KRAS mutant lung and colon cancer through transcriptional induction of ERBB3. Cell Rep. 7, 86–93 (2014).
Johnson, G. L., Stuhlmiller, T. J., Angus, S. P., Zawistowski, J. S. & Graves, L. M. Molecular pathways: adaptive kinome reprogramming in response to targeted inhibition of the BRAF-MEK-ERK pathway in cancer. Clin. Cancer Res. 20, 2516–2522 (2014).
Blumenschein, G. R. Jr et al. A randomized phase II study of the MEK1/MEK2 inhibitor trametinib (GSK1120212) compared with docetaxel in KRAS-mutant advanced non-small-cell lung cancer (NSCLC). Ann. Oncol. 26, 894–901 (2015).
Jänne, P. A. et al. Selumetinib plus docetaxel compared with docetaxel alone and progression-free survival in patients with KRAS-mutant advanced non-small cell lung cancer: the SELECT-1 randomized clinical trial. JAMA 317, 1844–1853 (2017).
Welsh, S. J. & Corrie, P. G. Management of BRAF and MEK inhibitor toxicities in patients with metastatic melanoma. Ther. Adv. Med. Oncol. 7, 122–136 (2015).
Duncan, J. S. et al. Dynamic reprogramming of the kinome in response to targeted MEK inhibition in triple-negative breast cancer. Cell 149, 307–321 (2012).
Zhao, Y. & Adjei, A. A. The clinical development of MEK inhibitors. Nat. Rev. Clin. Oncol. 11, 385–400 (2014).
Bollag, G. et al. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature 467, 596–599 (2010).
Morris, E. J. et al. Discovery of a novel ERK inhibitor with activity in models of acquired resistance to BRAF and MEK inhibitors. Cancer Discov. 3, 742–750 (2013).
Moschos, S. J. et al. Development of MK-8353, an orally administered ERK1/2 inhibitor, in patients with advanced solid tumors. JCI Insight 3, 92352 (2018).
Germann, U. A. et al. The selective ERK inhibitor BVD-523 is active in models of MAPK pathway-dependent cancers, including those with intrinsic and acquired drug resistance [abstract]. Cancer Res. 75 (Suppl), 4693 (2015).
Sullivan, R. J. et al. First-in-class ERK1/2 inhibitor ulixertinib (BVD-523) in patients with MAPK mutant advanced solid tumors: results of a phase I dose-escalation and expansion study. Cancer Discov. 8, 184–195 (2018).
Hatzivassiliou, G. et al. ERK inhibition overcomes acquired resistance to MEK inhibitors. Mol. Cancer Ther. 11, 1143–1154 (2012).
Hayes, T. K. et al. Long-term ERK inhibition in KRAS-mutant pancreatic cancer is associated with MYC degradation and senescence-like growth suppression. Cancer Cell 29, 75–89 (2016).
Goetz, E. M., Ghandi, M., Treacy, D. J., Wagle, N. & Garraway, L. A. ERK mutations confer resistance to mitogen-activated protein kinase pathway inhibitors. Cancer Res. 74, 7079–7089 (2014).
Jha, S. et al. Dissecting therapeutic resistance to ERK inhibition. Mol. Cancer Ther. 15, 548–559 (2016).
Brenan, L. et al. Phenotypic characterization of a comprehensive set of MAPK1/ERK2 missense mutants. Cell Rep. 17, 1171–1183 (2016).
Long, G. V. et al. Dabrafenib plus trametinib versus dabrafenib monotherapy in patients with metastatic BRAF V600E/K-mutant melanoma: long-term survival and safety analysis of a phase 3 study. Ann. Oncol. 28, 1631–1639 (2017).
Long, G. V. et al. Combined BRAF and MEK inhibition versus BRAF inhibition alone in melanoma. N. Engl. J. Med. 371, 1877–1888 (2014).
Ascierto, P. A. et al. Cobimetinib combined with vemurafenib in advanced BRAFV600-mutant melanoma (coBRIM): updated efficacy results from a randomised, double-blind, phase 3 trial. Lancet Oncol. 17, 1248–1260 (2016).
Hazar-Rethinam, M. et al. Convergent therapeutic strategies to overcome the heterogeneity of acquired resistance in BRAFV600E colorectal cancer. Cancer Discov. 8, 417–427 (2018).
Corcoran, R. B. et al. Combined BRAF, EGFR, and MEK inhibition in patients with BRAFV600E-mutant colorectal cancer. Cancer Discov. 8, 428–443 (2018).
Merchant, M. et al. Combined MEK and ERK inhibition overcomes therapy-mediated pathway reactivation in RAS mutant tumors. PLOS ONE 12, e0185862 (2017).
Vito, W. R. et al. Vertical inhibition of the MAPK pathway enhances therapeutic responses in NRAS-mutant melanoma. Pigment Cell Melanoma Res. 27, 1154–1158 (2014).
Vakana, E. et al. LY3009120, a panRAF inhibitor, has significant anti-tumor activity in BRAF and KRAS mutant preclinical models of colorectal cancer. Oncotarget 8, 9251–9266 (2017).
Hong, D. S. et al. A first-in-human dose phase 1 study of LY3009120 in advanced cancer patients. J. Clin. Oncol. 35, 2507–2507 (2017).
Girotti, M. R. et al. Paradox-breaking RAF inhibitors that also target SRC are effective in drug-resistant BRAF mutant melanoma. Cancer Cell 27, 85–96 (2015).
Wada, M., Horinaka, M., Yamazaki, T., Katoh, N. & Sakai, T. The dual RAF/MEK inhibitor CH5126766/RO5126766 may be a potential therapy for RAS-mutated tumor cells. PLOS ONE 9, e113217 (2014).
Martinez-Garcia, M. et al. First-in-human, phase I dose-escalation study of the safety, pharmacokinetics, and pharmacodynamics of RO5126766, a first-in-class dual MEK/RAF inhibitor in patients with solid tumors. Clin. Cancer Res. 18, 4806–4819 (2012).
Corcoran, R. B. et al. Synthetic lethal interaction of combined BCL-XL and MEK inhibition promotes tumor regressions in KRAS mutant cancer models. Cancer Cell 23, 121–128 (2013).
Okumura, S. & Jänne, P. A. Molecular pathways: the basis for rational combination using MEK inhibitors in KRAS-mutant cancers. Clin. Cancer Res. 20, 4193–4199 (2014).
Engelman, J. A. et al. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat. Med. 14, 1351–1356 (2008).
Posch, C. et al. Combined targeting of MEK and PI3K/mTOR effector pathways is necessary to effectively inhibit NRAS mutant melanoma in vitro and in vivo. Proc. Natl Acad. Sci. USA 110, 4015–4020 (2013).
Junttila, M. R. et al. Modeling targeted inhibition of MEK and PI3 kinase in human pancreatic cancer. Mol. Cancer Ther. 14, 40–47 (2015).
She, Q.-B. et al. 4E-BP1 is a key effector of the oncogenic activation of the AKT and ERK signaling pathways that integrates their function in tumors. Cancer Cell 18, 39–51 (2010).
Tolcher, A. W. et al. Antitumor activity in RAS-driven tumors by blocking AKT and MEK. Clin. Cancer Res. 21, 739–748 (2015).
Chung, V. et al. Effect of selumetinib and MK-2206 vs oxaliplatin and fluorouracil in patients with metastatic pancreatic cancer after prior therapy: SWOG S1115 study randomized clinical trial. JAMA Oncol. 3, 516–522 (2017).
Bedard, P. L. et al. A phase Ib dose-escalation study of the oral pan-PI3K inhibitor buparlisib (BKM120) in combination with the oral MEK1/2 inhibitor trametinib (GSK1120212) in patients with selected advanced solid tumors. Clin. Cancer Res. 21, 730–738 (2015).
Do, K. et al. Biomarker-driven phase 2 study of MK-2206 and selumetinib (AZD6244, ARRY-142886) in patients with colorectal cancer. Invest. New Drugs 33, 720–728 (2015).
Sun, C. et al. Rational combination therapy with PARP and MEK inhibitors capitalizes on therapeutic liabilities in RAS mutant cancers. Sci. Transl Med. 9, eaal5148 (2017).
Pek, M. et al. Oncogenic KRAS-associated gene signature defines co-targeting of CDK4/6 and MEK as a viable therapeutic strategy in colorectal cancer. Oncogene 36, 4975–4986 (2017).
Peeper, D. S. et al. Ras signalling linked to the cell-cycle machinery by the retinoblastoma protein. Nature 386, 177 (1997).
Chang, Z. et al. Cooperativity of oncogenic K-Ras and downregulated p16/INK4A in human pancreatic tumorigenesis. PLOS ONE 9, e101452 (2014).
Schuster, K. et al. Nullifying the CDKN2AB locus promotes mutant K-ras lung tumorigenesis. Mol. Cancer Res. 12, 912–923 (2014).
Sherr, C. J., Beach, D. & Shapiro, G. I. Targeting CDK4 and CDK6: from discovery to therapy. Cancer Discov. 6, 353–367 (2016).
Ziemke, E. K. et al. Sensitivity of KRAS-mutant colorectal cancers to combination therapy that cotargets MEK and CDK4/6. Clin. Cancer Res. 22, 405–414 (2016).
Franco, J., Witkiewicz, A. K. & Knudsen, E. S. CDK4/6 inhibitors have potent activity in combination with pathway selective therapeutic agents in models of pancreatic cancer. Oncotarget 5, 6512–6525 (2014).
Kwong, L. N. et al. Oncogenic NRAS signaling differentially regulates survival and proliferation in melanoma. Nat. Med. 18, 1503–1510 (2012).
Manchado, E. et al. A combinatorial strategy for treating KRAS-mutant lung cancer. Nature 534, 647–651 (2016).
Hata, A. N. et al. Failure to induce apoptosis via BCL-2 family proteins underlies lack of efficacy of combined MEK and PI3K inhibitors for KRAS-mutant lung cancers. Cancer Res. 74, 3146–3156 (2014).
Horn, T. et al. High-order drug combinations are required to effectively kill colorectal cancer cells. Cancer Res. 76, 6950–6963 (2016).
Lal, N. et al. KRAS mutation and consensus molecular subtypes 2 and 3 are independently associated with reduced immune infiltration and reactivity in colorectal cancer. Clin. Cancer Res. 24, 224–233 (2017).
Loi, S. et al. RAS/MAPK activation is associated with reduced tumor-infiltrating lymphocytes in triple-negative breast cancer: therapeutic cooperation between MEK and PD-1/PD-L1 immune checkpoint inhibitors. Clin. Cancer Res. 22, 1499–1509 (2016).
Pylayeva-Gupta, Y., Lee, K. E., Hajdu, C. H., Miller, G. & Bar-Sagi, D. Oncogenic Kras-induced GM-CSF production promotes the development of pancreatic neoplasia. Cancer Cell 21, 836–847 (2012).
Coelho, M. A. et al. Oncogenic RAS signaling promotes tumor immunoresistance by stabilizing PD-L1 mRNA. Immunity 47, 1083–1099 (2017).
Cullis, J., Das, S. & Bar-Sagi, D. Kras and tumor immunity: friend or foe? Cold Spring Harb. Perspect Med. 8, a031849 (2017).
Tran, E. et al. T-cell transfer therapy targeting mutant KRAS in cancer. N. Engl. J. Med. 375, 2255–2262 (2016).
June, C. H., Riddell, S. R. & Schumacher, T. N. Adoptive cellular therapy: a race to the finish line. Sci. Transl Med. 7, 280ps7 (2015).
June, C. H. Drugging the undruggable Ras — immunotherapy to the rescue? N. Engl. J. Med. 375, 2286–2289 (2016).
Wang, Q. J. et al. Identification of T cell receptors targeting KRAS-mutated human tumors. Cancer Immunol. Res. 4, 204–214 (2016).
Eriksen, J. A., Gladhaug, I. P., Rosseland, A., Risberg Handeland, K. & Buanes, T. An observational clinical study with RAS peptide vaccine TG01 evaluating immune response, safety and overall survival in patients with non-resectable pancreatic cancer. Ann. Oncol. 28, mdx376.018 (2017).
Palmer, D. H., Dueland, S., Valle, J. W. & Aksnes, A.-K. A phase I/II trial of TG01/GM-CSF and gemcitabine as adjuvant therapy for treating patients with resected RAS-mutant adenocarcinoma of the pancreas. J. Clin. Oncol. 35, 4119–4119 (2017).
Topalian Suzanne, L., Drake Charles, G. & Pardoll Drew, M. Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell 27, 450–461 (2015).
Cooper, Z. A. et al. Response to BRAF inhibition in melanoma is enhanced when combined with immune checkpoint blockade. Cancer Immunol. Res. 2, 643–654 (2014).
Ebert, P. J. R. et al. MAP kinase inhibition promotes T cell and anti-tumor activity in combination with PD-L1 checkpoint blockade. Immunity 44, 609–621 (2016).
Bendell, J. C. et al. Clinical activity and safety of cobimetinib (cobi) and atezolizumab in colorectal cancer (CRC). J. Clin. Oncol. 34, 3502–3502 (2016).
Le, D. T. et al. PD-1 blockade in tumors with mismatch-repair deficiency. N. Engl. J. Med. 372, 2509–2520 (2015).
Infante, J. R. et al. Safety, pharmacokinetic, pharmacodynamic, and efficacy data for the oral MEK inhibitor trametinib: a phase 1 dose-escalation trial. Lancet Oncol. 13, 773–781 (2012).
Bendell, J. C. et al. A phase Ib study of safety and clinical activity of atezolizumab (A) and cobimetinib (C) in patients (pts) with metastatic colorectal cancer (mCRC). J. Clin. Oncol. 36, 560–560 (2018).
Acknowledgements
The work of R.B.C. is supported by US National Institutes of Health (NIH)-National Cancer Institute (NCI) Gastrointestinal Cancer SPORE P50 CA127003, R01CA208437 and U54CA224068 and a Stand Up To Cancer (SU2C) Colorectal Dream Team Translational Research Grant (grant number SU2C-AACR-DT22-17). Research grants are administered by the American Association of Cancer Research, the scientific partner of SU2C.
Author information
Authors and Affiliations
Contributions
M.B.R. and R.B.C. contributed equally to all aspects of the preparation of this manuscript.
Corresponding author
Ethics declarations
Competing interests
R.B.C. has received consulting fees from Amgen, Astex, Avidity, Bristol-Myers Squibb, FOG Pharma, Genentech, LOXO, Merrimack, N-of-One, Roche, Roivant, Shire, Symphogen, Taiho and Warp Drive Bio, as well as research support from AstraZeneca and Sanofi. M.B.R. declares no competing interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
The RAS initiative: https://www.cancer.gov/research/key-initiatives/rasUS National Institutes of Health ClinicalTrials.gov database: https://www.clinicaltrials.gov
Rights and permissions
About this article
Cite this article
Ryan, M.B., Corcoran, R.B. Therapeutic strategies to target RAS-mutant cancers. Nat Rev Clin Oncol 15, 709–720 (2018). https://doi.org/10.1038/s41571-018-0105-0
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41571-018-0105-0
This article is cited by
-
Harnessing ferroptosis for enhanced sarcoma treatment: mechanisms, progress and prospects
Experimental Hematology & Oncology (2024)
-
Combinatorial strategies to target RAS-driven cancers
Nature Reviews Cancer (2024)
-
Targeting KRAS in cancer
Nature Medicine (2024)
-
CDK4/6 inhibition sensitizes MEK inhibition by inhibiting cell cycle and proliferation in pancreatic ductal adenocarcinoma
Scientific Reports (2024)
-
The role of mitochondria in the resistance of melanoma to PD-1 inhibitors
Journal of Translational Medicine (2023)