Abstract
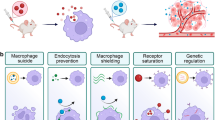
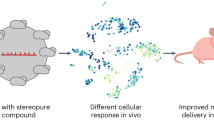
Nanomedicines have been approved to treat multiple human diseases. However, clinical adoption of nanoformulated agents is often hindered by concerns about hepatic uptake and clearance, a process that is not fully understood. Here we show that the antitumour efficacy of cancer nanomedicine exhibits an age-associated disparity. Tumour delivery and treatment outcomes are superior in old versus young mice, probably due to an age-related decline in the ability of hepatic phagocytes to take up and remove nanoparticles. Transcriptomic- and protein-level analysis at the single-cell and bulk levels reveals an age-associated decrease in the numbers of hepatic macrophages that express the scavenger receptor MARCO in mice, non-human primates and humans. Therapeutic blockade of MARCO is shown to decrease the phagocytic uptake of nanoparticles and improve the antitumour effect of clinically approved cancer nanotherapeutics in young but not aged mice. Together, these results reveal an age-associated disparity in the phagocytic clearance of nanotherapeutics that affects their antitumour response, thus providing a strong rationale for an age-appropriate approach to cancer nanomedicine.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All data supporting the findings of this study are provided in the figures and supplementary materials. The sequencing data have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus database under accession number GSE205833. Human liver transcriptome data were obtained from the Human Protein Atlas v.20.1. Statistical source data for all figures and supplementary figures are provided with this paper. Additional information can be requested from the corresponding authors (W.J., B.Y.S.K., H.W.). All equipment and reagents are commercially available and are described in the Methods section. Source data are provided with this paper.
Code availability
All the codes used in this study are open source and are accessible to the public. The R packages used are indicated in the Methods.
References
de Magalhaes, J. P. How ageing processes influence cancer. Nat. Rev. Cancer 13, 357–365 (2013).
Laconi, E., Marongiu, F. & DeGregori, J. Cancer as a disease of old age: changing mutational and microenvironmental landscapes. Br. J. Cancer 122, 943–952 (2020).
Van Herck, Y. et al. Is cancer biology different in older patients? Lancet Healthy Longev. 2, E663–E677 (2021).
Sceneay, J. et al. Interferon signaling is diminished with age and is associated with immune checkpoint blockade efficacy in triple-negative breast cancer. Cancer Discov. 9, 1208–1227 (2019).
Kaur, A. et al. sFRP2 in the aged microenvironment drives melanoma metastasis and therapy resistance. Nature 532, 250–254 (2016).
Kim, B. Y., Rutka, J. T. & Chan, W. C. Nanomedicine. N. Engl. J. Med. 363, 2434–2443 (2010).
Jiang, W. et al. Designing nanomedicine for immuno-oncology. Nat. Biomed. Eng 1, 0029 (2017).
Rodriguez, P. L. et al. Minimal ‘self’ peptides that inhibit phagocytic clearance and enhance delivery of nanoparticles. Science 339, 971–975 (2013).
Parodi, A. et al. Synthetic nanoparticles functionalized with biomimetic leukocyte membranes possess cell-like functions. Nat. Nanotechnol. 8, 61–68 (2013).
Gradishar, W. J. et al. Phase III trial of nanoparticle albumin-bound paclitaxel compared with polyethylated castor oil-based paclitaxel in women with breast cancer. J. Clin. Oncol. 23, 7794–7803 (2005).
Barenholz, Y. Doxil®—the first FDA-approved nano-drug: lessons learned. J. Control. Release 160, 117–134 (2012).
Mitchell, M. J. et al. Engineering precision nanoparticles for drug delivery. Nat. Rev. Drug Discov. 20, 101–124 (2021).
Blanco, E., Shen, H. & Ferrari, M. Principles of nanoparticle design for overcoming biological barriers to drug delivery. Nat. Biotechnol. 33, 941–951 (2015).
Ngo, W. et al. Identifying cell receptors for the nanoparticle protein corona using genome screens. Nat. Chem. Biol. 18, 1023–1031 (2022).
Boehnke, N. et al. Massively parallel pooled screening reveals genomic determinants of nanoparticle delivery. Science 377, eabm5551 (2022).
Dobrowolski, C. et al. Nanoparticle single-cell multiomic readouts reveal that cell heterogeneity influences lipid nanoparticle-mediated messenger RNA delivery. Nat. Nanotechnol. 17, 871–879 (2022).
Jiang, W., Wang, Y., Wargo, J. A., Lang, F. F. & Kim, B. Y. S. Considerations for designing preclinical cancer immune nanomedicine studies. Nat. Nanotechnol. 16, 6–15 (2021).
Ouyang, B. et al. The dose threshold for nanoparticle tumour delivery. Nat. Mater. 19, 1362–1371 (2020).
Wilhelm, S. et al. Analysis of nanoparticle delivery to tumours. Nat. Rev. Mater. 1, 1–12 (2016).
Chen, Y. et al. Therapeutic remodeling of the tumor microenvironment enhances nanoparticle delivery. Adv. Sci. (Weinh.) 6, 1802070 (2019).
Pili, R. et al. Altered angiogenesis underlying age-dependent changes in tumor growth. J. Natl Cancer Inst. 86, 1303–1314 (1994).
Marinho, A., Soares, R., Ferro, J., Lacerda, M. & Schmitt, F. C. Angiogenesis in breast cancer is related to age but not to other prognostic parameters. Pathol. Res. Pract. 193, 267–273 (1997).
Ouyang, B. et al. Impact of tumor barriers on nanoparticle delivery to macrophages. Mol. Pharm. 19, 1917–1925 (2022).
Grolleau, A., Misek, D. E., Kuick, R., Hanash, S. & Mule, J. J. Inducible expression of macrophage receptor Marco by dendritic cells following phagocytic uptake of dead cells uncovered by oligonucleotide arrays. J. Immunol. 171, 2879–2888 (2003).
Hamilton, R. F. Jr., Thakur, S. A., Mayfair, J. K. & Holian, A. MARCO mediates silica uptake and toxicity in alveolar macrophages from C57BL/6 mice. J. Biol. Chem. 281, 34218–34226 (2006).
Park, J. et al. Intravascular innate immune cells reprogrammed via intravenous nanoparticles to promote functional recovery after spinal cord injury. Proc. Natl Acad. Sci. USA 116, 14947–14954 (2019).
Pikkarainen, T., Brannstrom, A. & Tryggvason, K. Expression of macrophage MARCO receptor induces formation of dendritic plasma membrane processes. J. Biol. Chem. 274, 10975–10982 (1999).
Hirano, S., Fujitani, Y., Furuyama, A. & Kanno, S. Macrophage receptor with collagenous structure (MARCO) is a dynamic adhesive molecule that enhances uptake of carbon nanotubes by CHO-K1 cells. Toxicol. Appl Pharm. 259, 96–103 (2012).
van der Laan, L. J. et al. Regulation and functional involvement of macrophage scavenger receptor MARCO in clearance of bacteria in vivo. J. Immunol. 162, 939–947 (1999).
Arredouani, M. S. et al. MARCO is the major binding receptor for unopsonized particles and bacteria on human alveolar macrophages. J. Immunol. 175, 6058–6064 (2005).
Li, Z. et al. Aging-impaired filamentous actin polymerization signaling reduces alveolar macrophage phagocytosis of bacteria. J. Immunol. 199, 3176–3186 (2017).
Ojala, J. R., Pikkarainen, T., Tuuttila, A., Sandalova, T. & Tryggvason, K. Crystal structure of the cysteine-rich domain of scavenger receptor MARCO reveals the presence of a basic and an acidic cluster that both contribute to ligand recognition. J. Biol. Chem. 282, 16654–16666 (2007).
Novakowski, K. E. et al. A naturally occurring transcript variant of MARCO reveals the SRCR domain is critical for function. Immunol. Cell Biol. 94, 646–655 (2016).
Brannstrom, A., Sankala, M., Tryggvason, K. & Pikkarainen, T. Arginine residues in domain V have a central role for bacteria-binding activity of macrophage scavenger receptor MARCO. Biochem. Biophys. Res. Commun. 290, 1462–1469 (2002).
Wang, Y. et al. Mutant LKB1 confers enhanced radiosensitization in combination with trametinib in KRAS-mutant non-small cell lung cancer. Clin. Cancer Res. 24, 5744–5756 (2018).
Guilliams, M. et al. Spatial proteogenomics reveals distinct and evolutionarily conserved hepatic macrophage niches. Cell 185, 379–396 (2022).
Tabula Sapiens Consortium. The Tabula Sapiens: a multiple-organ, single-cell transcriptomic atlas of humans. Science 376, eabl4896 (2022).
Uhlen, M. et al. Proteomics. Tissue-based map of the human proteome. Science 347, 1260419 (2015).
Acknowledgements
The authors thank C. F. Wogan of the Division of Radiation Oncology at MD Anderson Cancer Center for editing this manuscript. The authors thank B. Q. Tran and S. A. Martinez at MD Anderson’s Metabolomics Facility for assisting with the mass spectrometry experiment and data analysis; J. Zhang of MD Anderson’s Department of Experimental Radiation Oncology for processing histologic samples; V. Van and K. L. Maldonado at MD Anderson’s Small Animal Imaging Facility for helping with the animal experiments; N. R. Vaughn and N. Nguyen at MD Anderson’s Flow Cytometry and Cellular Imaging Core Facility for helping with flow cytometry experiments; E. Parks at the UT Southwestern Medical Center for performing some of the in vitro experiments; and C. Yang at Washington University in St Louis for helping with bulk RNA-seq analysis. This work was supported in part by the Cancer Prevention and Research Institute of Texas (CPRIT) (RR180017, W.J.) and Radiation Oncology Institute (N.N.S. and W.J.). This work was also supported in part by National Institutes of Health grant P30CA016672 (principal investigator, P. Pisters).
Author information
Authors and Affiliations
Contributions
W.J., B.Y.S.K., H.W. and Y.W. conceived the project and were responsible for all phases of the research. Y.W., W.D., D.L., L.Y., R.Y., H.W., B.Y.S.K. and W.J. designed the experiments. Y.W., D.L., L.Y., X.L., Y.L., M.Y., A.A., P.G., S.D., K.H., J.H., R.Y., L.T. and F.Z. performed the experiments and collected the data. W.D., L.Y. and Y.Z. performed bioinformatics and statistical analysis. Y.W., W.D., L.Y., X.L., Z.Y., M.Y., P.G., S.D., R.Y., K.H., B.R.S., M.K., Y.L., L.T., P.L.L., T.D.G., S.K.T., Y.Z., J.L., N.N.S. and W.J. analysed and interpreted the data. The manuscript was drafted by Y.W., W.D., L.Y., H.W., B.Y.S.K. and W.J. and was revised and approved by all authors.
Corresponding authors
Ethics declarations
Competing interests
A provisional patent application based on the technology described in the manuscript has been filed by the Board of Regents, The University of Texas System, with W.J., Y.W. and B.Y.S.K. as inventors. The other authors declare no competing interests.
Peer review
Peer review information
Nature Nanotechnology thanks Dennis Discher and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–51 and Table 1.
Supplementary Data
Statistical source data of supplementary figures and differentially expressed genes identified in single-cell sequencing
Source data
Source Data Figs. 1–5
Statistical Source data
Source Data Fig. 4
Unprocessed blots
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, Y., Deng, W., Lee, D. et al. Age-associated disparity in phagocytic clearance affects the efficacy of cancer nanotherapeutics. Nat. Nanotechnol. 19, 255–263 (2024). https://doi.org/10.1038/s41565-023-01502-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41565-023-01502-3