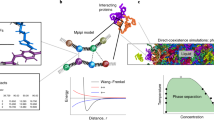
Abstract
Understanding the relationship between a polypeptide sequence and its phase separation has important implications for analysing cellular function, treating disease and designing novel biomaterials. Several sequence features have been identified as drivers for protein liquid–liquid phase separation (LLPS), schematized as a ‘molecular grammar’ for LLPS. Here we further probe how sequence modulates phase separation and the material properties of the resulting condensates, targeting sequence features previously overlooked in the literature. We generate sequence variants of a repeat polypeptide with either no charged residues, high net charge, no glycine residues or devoid of aromatic or arginine residues. All but one of 12 variants exhibited LLPS, albeit to different extents, despite substantial differences in composition. Furthermore, we find that all the condensates formed behaved like viscous fluids, despite large differences in their viscosities. Our results support the model of multiple interactions between diverse residue pairs—not just a handful of residues—working in tandem to drive the phase separation and dynamics of condensates.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Starting and ending configurations for all atomistic simulations have been deposited on Zenodo (https://doi.org/10.5281/zenodo.10523201). Materials are available upon reasonable request to the corresponding authors. Source data are provided with this paper.
Code availability
Codes to run and analyse atomistic simulations are available publicly and can be found at https://openmm.org/, https://gromacs.org/ and https://www.mdanalysis.org/. Codes to reproduce residue pairwise contacts and angle-versus-distance distributions have been deposited on Zenodo (https://doi.org/10.5281/zenodo.10523201).
References
Feric, M. et al. Coexisting liquid phases underlie nucleolar subcompartments. Cell 165, 1686–1697 (2016).
Brangwynne, C. P. et al. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 324, 1729–1732 (2009).
Lyon, A. S., Peeples, W. B. & Rosen, M. K. A framework for understanding the functions of biomolecular condensates across scales. Nat. Rev. Mol. Cell Biol. 22, 215–235 (2021).
Alberti, S. & Dormann, D. Liquid–liquid phase separation in disease. Annu. Rev. Genet. 53, 171–194 (2019).
Alberti, S. & Hyman, A. A. Biomolecular condensates at the nexus of cellular stress, protein aggregation disease and ageing. Nat. Rev. Mol. Cell Biol. 22, 196–213 (2021).
Shin, Y. & Brangwynne, C. P. Liquid phase condensation in cell physiology and disease. Science 357, eaaf4382 (2017).
Martin, E. W. & Holehouse, A. S. Intrinsically disordered protein regions and phase separation: sequence determinants of assembly or lack thereof. Emerg. Top. Life Sci. 4, 307–329 (2020).
Schuster, B. S. et al. Biomolecular condensates: sequence determinants of phase separation, microstructural organization, enzymatic activity and material properties. J. Phys. Chem. B 125, 3441–3451 (2021).
Borcherds, W., Bremer, A., Borgia, M. B. & Mittag, T. How do intrinsically disordered protein regions encode a driving force for liquid–liquid phase separation? Curr. Opin. Struct. Biol. 67, 41–50 (2021).
Dignon, G. L., Best, R. B. & Mittal, J. Biomolecular phase separation: from molecular driving forces to macroscopic properties. Annu. Rev. Phys. Chem. 71, 53–75 (2020).
Quiroz, F. G. & Chilkoti, A. Sequence heuristics to encode phase behaviour in intrinsically disordered protein polymers. Nat. Mater. 14, 1164–1171 (2015).
Dzuricky, M., Rogers, B. A., Shahid, A., Cremer, P. S. & Chilkoti, A. De novo engineering of intracellular condensates using artificial disordered proteins. Nat. Chem. 12, 814–825 (2020).
Wang, B., Patkar, S. S. & Kiick, K. L. Application of thermoresponsive intrinsically disordered protein polymers in nanostructured and microstructured materials. Macromol. Biosci. 21, 2100129 (2021).
Garcia Garcia, C., Patkar, S. S., Jovic, N., Mittal, J. & Kiick, K. L. Alteration of microstructure in biopolymeric hydrogels via compositional modification of resilin-like polypeptides. ACS Biomater. Sci. Eng. 7, 4244–4257 (2021).
Good, M. C., Zalatan, J. G. & Lim, W. A. Scaffold proteins: hubs for controlling the flow of cellular information. Science 332, 680–686 (2011).
Schuster, B. S. et al. Controllable protein phase separation and modular recruitment to form responsive membraneless organelles. Nat. Commun. 9, 2985 (2018).
Simon, J. R., Eghtesadi, S. A., Dzuricky, M., You, L. & Chilkoti, A. Engineered ribonucleoprotein granules inhibit translation in protocells. Mol. Cell 75, 66–75 (2019).
Nott, T. J. et al. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol. Cell 57, 936–947 (2015).
Brady, J. P. et al. Structural and hydrodynamic properties of an intrinsically disordered region of a germ cell-specific protein on phase separation. Proc. Natl Acad. Sci. USA 114, E8194–E8203 (2017).
Vernon, R. M. et al. Pi-Pi contacts are an overlooked protein feature relevant to phase separation. eLife 7, e31486 (2018).
Wang, J. et al. A molecular grammar governing the driving forces for phase separation of prion-like RNA binding proteins. Cell 174, 688–699 (2018).
Murthy, A. C. et al. Molecular interactions underlying liquid–liquid phase separation of the FUS low-complexity domain. Nat. Struct. Mol. Biol. 26, 637–648 (2019).
Martin, E. W. et al. Valence and patterning of aromatic residues determine the phase behavior of prion-like domains. Science 367, 694–699 (2020).
Schuster, B. S. et al. Identifying sequence perturbations to an intrinsically disordered protein that determine its phase-separation behavior. Proc. Natl Acad. Sci. USA 117, 11421–11431 (2020).
Murthy, A. C. et al. Molecular interactions contributing to FUS SYGQ LC-RGG phase separation and co-partitioning with RNA polymerase II heptads. Nat. Struct. Mol. Biol. 28, 923–935 (2021).
Bremer, A. et al. Deciphering how naturally occurring sequence features impact the phase behaviours of disordered prion-like domains. Nat. Chem. 14, 196–207 (2022).
Martin, E. W. & Mittag, T. Relationship of sequence and phase separation in protein low-complexity regions. Biochemistry 57, 2478–2487 (2018).
Tompa, P., Schad, E., Tantos, A. & Kalmar, L. Intrinsically disordered proteins: emerging interaction specialists. Curr. Opin. Struct. Biol. 35, 49–59 (2015).
Lin, Y., Currie, S. L. & Rosen, M. K. Intrinsically disordered sequences enable modulation of protein phase separation through distributed tyrosine motifs. J. Biol. Chem. 292, 19110–19120 (2017).
Holehouse, A. S., Ginell, G. M., Griffith, D. & Boke, E. Clustering of aromatic residues in prion-like domains can tune the formation, state and organization of biomolecular condensates: published as part of the Biochemistry virtual special issue ‘Protein condensates’. Biochemistry 60, 3566–3581 (2021).
Fisher, R. S. & Elbaum-Garfinkle, S. Tunable multiphase dynamics of arginine and lysine liquid condensates. Nat. Commun. 11, 4628 (2020).
Devarajan, D. S. et al. Effect of charge distribution on the dynamics of polyampholytic disordered proteins. Macromolecules 55, 8987–8997 (2022).
Rekhi, S. et al. Role of strong localized vs weak distributed interactions in disordered protein phase separation. J. Phys. Chem. B 127, 3829–3838 (2023).
Dai, Y. et al. Programmable synthetic biomolecular condensates for cellular control. Nat. Chem. Biol. 19, 518–528 (2023).
Cai, H., Vernon, R. M. & Forman-Kay, J. D. An interpretable machine-learning algorithm to predict disordered protein phase separation based on biophysical interactions. Biomolecules 12, 1131 (2022).
Hardenberg, M., Horvath, A., Ambrus, V., Fuxreiter, M. & Vendruscolo, M. Widespread occurrence of the droplet state of proteins in the human proteome. Proc. Natl Acad. Sci. USA 117, 33254–33262 (2020).
Bolognesi, B. et al. A concentration-dependent liquid phase separation can cause toxicity upon increased protein expression. Cell Rep. 16, 222–231 (2016).
Mohanty, P. et al. Principles governing the phase separation of multidomain proteins. Biochemistry 61, 2443–2455 (2022).
Zheng, W. et al. Molecular details of protein condensates probed by microsecond long atomistic simulations. J. Phys. Chem. B 124, 11671–11679 (2020).
Kim, S. et al. Complexation and coacervation of like-charged polyelectrolytes inspired by mussels. Proc. Natl Acad. Sci. USA 113, E847–E853 (2016).
Baruch Leshem, A. et al. Biomolecular condensates formed by designer minimalistic peptides. Nat. Commun. 14, 421 (2023).
Van Treeck, B. et al. RNA self-assembly contributes to stress granule formation and defining the stress granule transcriptome. Proc. Natl Acad. Sci. USA 115, 2734–2739 (2018).
Paloni, M., Bailly, R., Ciandrini, L. & Barducci, A. Unraveling molecular interactions in liquid-liquid phase separation of disordered proteins by atomistic simulations. J. Phys. Chem. B 124, 9009–9016 (2020).
Armstrong, C. T., Mason, P. E., Anderson, J. R. & Dempsey, C. E. Arginine side chain interactions and the role of arginine as a gating charge carrier in voltage sensitive ion channels. Sci. Rep. 6, 21759 (2016).
Lee, D., Lee, J. & Seok, C. What stabilizes close arginine pairing in proteins? Phys. Chem. Chem. Phys. 15, 5844–5853 (2013).
Harms, M. J., Schlessman, J. L., Sue, G. R. & García-Moreno, B. Arginine residues at internal positions in a protein are always charged. Proc. Natl Acad. Sci. USA 108, 18954–18959 (2011).
Ray, S. et al. α-Synuclein aggregation nucleates through liquid–liquid phase separation. Nat. Chem. 12, 705–716 (2020).
Alshareedah, I., Moosa, M. M., Pham, M., Potoyan, D. A. & Banerjee, P. R. Programmable viscoelasticity in protein–RNA condensates with disordered sticker-spacer polypeptides. Nat. Commun. 12, 6620 (2021).
Jawerth, L. et al. Protein condensates as aging Maxwell fluids. Science 370, 1317–1323 (2020).
Sundaravadivelu Devarajan, D. et al. Sequence-dependent material properties of biomolecular condensates and their relation to dilute phase conformations. Nat. Commun. 15, 1912 (2024).
An, Y., Webb, M. A. & Jacobs, W. M. Active learning of the thermodynamics-dynamics trade-off in protein condensates. Sci. Adv. 10, eadj2448 (2024).
Urry, D. W. et al. Hydrophobicity scale for proteins based on inverse temperature transitions. Biopolymers 32, 1243–1250 (1992).
Zeng, X. & Pappu, R. V. Developments in describing equilibrium phase transitions of multivalent associative macromolecules. Curr. Opin. Struct. Biol. 79, 102540 (2023).
Pappu, R. V., Cohen, S. R., Dar, F., Farag, M. & Kar, M. Phase transitions of associative biomacromolecules. Chem. Rev. 123, 8945–8987 (2023).
Dill, K. & Bromberg, S. Molecular Driving Forces: Statistical Thermodynamics in Biology, Chemistry, Physics and Nanoscience (Garland Science, 2010).
Yang, Y., Jones, H. B., Dao, T. P. & Castañeda, C. A. Single amino acid substitutions in stickers, but not spacers, substantially alter UBQLN2 phase transitions and dense phase material properties. J. Phys. Chem. B 123, 3618–3629 (2019).
König, B., Pezzotti, S., Ramos, S., Schwaab, G. & Havenith, M. Real-time measure of solvation free energy changes upon liquid-liquid phase separation of α-elastin. Biophys. J. https://doi.org/10.1016/j.bpj.2023.07.023 (2023).
Pezzotti, S., König, B., Ramos, S., Schwaab, G. & Havenith, M. Liquid-liquid phase separation? Ask the water! J. Phys. Chem. Lett. 14, 1556–1563 (2023).
Wolfenden, R., Andersson, L., Cullis, P. & Southgate, C. Affinities of amino acid side chains for solvent water. Biochemistry 20, 849–855 (1981).
Chu, X. et al. Prediction of liquid-liquid phase separating proteins using machine learning. BMC Bioinformatics 23, 72 (2022).
Saar, K. L. et al. Learning the molecular grammar of protein condensates from sequence determinants and embeddings. Proc. Natl Acad. Sci. USA 118, e2019053118 (2021).
Ng, S. C. & Görlich, D. A simple thermodynamic description of phase separation of Nup98 FG domains. Nat. Commun. 13, 6172 (2022).
Gallivan, J. P. & Dougherty, D. A. Cation-π interactions in structural biology. Proc. Natl Acad. Sci. USA 96, 9459–9464 (1999).
Kumar, K. et al. Cation-π interactions in protein-ligand binding: theory and data-mining reveal different roles for lysine and arginine. Chem. Sci. 9, 2655–2665 (2018).
Sabari, B. R. et al. Coactivator condensation at super-enhancers links phase separation and gene control. Science 361, eaar3958 (2018).
Flory, P. J. Thermodynamics of high polymer solutions. J. Chem. Phys. 10, 51–61 (1942).
Lin, Y.-H., Song, J., Forman-Kay, J. D. & Chan, H. S. Random-phase-approximation theory for sequence-dependent, biologically functional liquid-liquid phase separation of intrinsically disordered proteins. J. Mol. Liq. 228, 176–193 (2017).
Charati, M. B., Ifkovits, J. L., Burdick, J. A., Linhardt, J. G. & Kiick, K. L. Hydrophilic elastomeric biomaterials based on resilin-like polypeptides. Soft Matter 5, 3412–3416 (2009).
Li, L., Teller, S., Clifton, R. J., Jia, X. & Kiick, K. L. Tunable mechanical stability and deformation response of a resilin-based elastomer. Biomacromolecules 12, 2302–2310 (2011).
Li, L., Tong, Z., Jia, X. & Kiick, K. L. Resilin-like polypeptide hydrogels engineered for versatile biological function. Soft Matter 9, 665–673 (2013).
Li, L. & Kiick, K. L. Transient dynamic mechanical properties of resilin-based elastomeric hydrogels. Front. Chem. 2, 21 (2014).
Tsolis, A. C., Papandreou, N. C., Iconomidou, V. A. & Hamodrakas, S. J. A consensus method for the prediction of ‘aggregation-prone’ peptides in globular proteins. PLoS One 8, e54175 (2013).
Allan, D. B., Thomas, C., Keim, N. C., van der Wel, C. M. & Verweij, R. W. soft-matter/trackpy: Trackpy v0.5.0 (Zenodo, 2021); https://doi.org/10.5281/zenodo.4682814
Regy, R. M., Thompson, J., Kim, Y. C. & Mittal, J. Improved coarse‐grained model for studying sequence dependent phase separation of disordered proteins. Protein Sci. 30, 1371–1379 (2021).
Webb, B. & Sali, A. Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinformatics 54, 5.6.1–5.6.37 (2016).
Vitalis, A. & Pappu, R. V. Methods for Monte Carlo simulations of biomacromolecules. Annu. Rep. Comput. Chem. 5, 49–76 (2009).
Vitalis, A. & Pappu, R. V. ABSINTH: a new continuum solvation model for simulations of polypeptides in aqueous solutions. J. Comput. Chem. 30, 673–699 (2009).
Tang, W. S., Fawzi, N. L. & Mittal, J. Refining all-atom protein force fields for polar-rich, prion-like, low-complexity intrinsically disordered proteins. J. Phys. Chem. B 124, 9505–9512 (2020).
Abascal, J. L. & Vega, C. A general purpose model for the condensed phases of water: TIP4P/2005. J. Chem. Phys. 123, 234505 (2005).
Luo, Y. & Roux, B. Simulation of osmotic pressure in concentrated aqueous salt solutions. J. Phys. Chem. Lett. 1, 183–189 (2010).
Abraham, M. J. et al. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1, 19–25 (2015).
Evans, D. J. & Holian, B. L. The nose-hoover thermostat. J. Chem. Phys. 83, 4069–4074 (1985).
Berendsen, H. J., Postma, J. V., Van Gunsteren, W. F., DiNola, A. & Haak, J. R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 81, 3684–3690 (1984).
Zhang, Z., Liu, X., Yan, K., Tuckerman, M. E. & Liu, J. Unified efficient thermostat scheme for the canonical ensemble with holonomic or isokinetic constraints via molecular dynamics. J. Phys. Chem. A 123, 6056–6079 (2019).
Eastman, P. et al. OpenMM 7: rapid development of high performance algorithms for molecular dynamics. PLoS Comput. Biol. 13, e1005659 (2017).
Darden, T., York, D. & Pedersen, L. Particle mesh Ewald: an N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 98, 10089–10092 (1993).
Ryckaert, J.-P., Ciccotti, G. & Berendsen, H. J. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 23, 327–341 (1977).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Michaud‐Agrawal, N., Denning, E. J., Woolf, T. B. & Beckstein, O. MDAnalysis: a toolkit for the analysis of molecular dynamics simulations. J. Comput. Chem. 32, 2319–2327 (2011).
Gowers, R. J. et al. MDAnalysis:A Python package for the rapid analysis of molecular dynamics simulations. In Proc. 15th Python in Science Conference (eds Benthall, S. & Rostrup, S.) 98 (SciPy, 2016).
Case, D. A. et al. Amber 2021 (Univ. California San Francisco, 2021).
Galvanetto, N. et al. Extreme dynamics in a biomolecular condensate. Nature 619, 876–883 (2023).
Humphrey, W., Dalke, A. & Schulten, K. VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–38 (1996).
Pettersen, E. F. et al. UCSF ChimeraX: structure visualization for researchers, educators and developers. Protein Sci. 30, 70–82 (2021).
Li, W. et al. Biophysical properties of AKAP95 protein condensates regulate splicing and tumorigenesis. Nat. Cell Biol. 22, 960–972 (2020).
Conicella, A. E. et al. TDP-43 α-helical structure tunes liquid-liquid phase separation and function. Proc. Natl Acad. Sci. USA 117, 5883–5894 (2020).
Acknowledgements
This Article is based on research supported in part by the National Science Foundation (DMR-2004796 to K.L.K. and J.M.), the National Institute of General Medical Science of the National Institute of Health (R01GM136917 to J.M. and R35GM142903 to B.S.S.) and the Welch Foundation (A-2113-202203311 to J.M.). Use of the Texas A&M High Performance Research Computing is greatly acknowledged for the computational resources utilized in this work. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
S.R., C.G.G., K.L.K. and J.M. conceived the research. B.S.S., K.L.K. and J.M. designed and supervised the research. S.R. and J.M. designed the sequences. C.G.G. expressed and purified all polypeptides and performed turbidity experiments. M.B. performed microscopy, microrheology and additional turbidity experiments. S.R. performed the simulations. S.R. analysed the simulations aided by A.R. S.R., C.G.G., M.B., B.S.S. and J.M. wrote the paper with help from the other authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Chemistry thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Salt dependence of polycation LLPS.
(a) Turbidity curves for the polycationic GRGNSPYS variant at three salt concentrations (50, 137, and 300 mM NaCl). Three independent experiments are shown at each concentration. (b) Transition temperatures estimated from the turbidity curves as shown in (a). Data are presented as mean values +/− SD, n = 3 independent experiments.
Extended Data Fig. 2 Pairwise contacts of R-to-Q and D-to-N variants.
Average residue pairwise contacts for the (a) GRGNSPYS and (b) GQGDSPYS variants with respect to WT. Residue pairs not involving mutated residues are shown as dark gray circles while residue pairs involving the mutated residues are shown as red squares (D-to-N) or orange diamonds (R-to-Q).
Extended Data Fig. 3 Turbidity and partial phase diagram of GRGASPYA.
Turbidity (a) and partial phase diagram (b) of GRGASPYA at different concentrations of protein in PBS. Data are presented as mean values +/− SD, n = 3 independent experiments.
Extended Data Fig. 4 Pairwise contacts of aromatic substitutions in the polycation.
Average residue pairwise contacts for the (a) GRGNSPFS and (b) GRGNSPWS variants with respect to GRGNSPYS. Upper plots: Residue pairs not involving mutated residues are shown as dark gray circles while residue pairs involving the mutated residues are shown as purple triangles. Lower plots: Contact ratio between residue pairs for the GRGNSPFS and GRGNSPWS variants to that of the GRGNSPYS variant.
Extended Data Fig. 5 Material properties of A-IDP condensates.
(a) FRAP of WT and GRGNSPWS with RGG-GFP-RGG as a fluorescent tracer. Data are presented as mean values +/− SD, n = 4 (WT), n = 9 (GRGNSPWS) different condensates from one experiment. (b) Effect of total protein concentration on condensate viscosity, as measured by microrheology. Measurements were conducted for GRGNSPFS and GQGNSPYS, at two concentrations each. Data are presented as mean values +/− SD, n = 4 videos from one experiment. (c) MSDs measured in polycationic GRGNSPYS condensates using PEGylated and carboxylated beads. (d) Viscosity of GRGDSPYS (WT), GQGNSPYS, and GRGNSPYS determined by particle tracking microrheology of 0.5 μm PEGylated vs. carboxylated beads. Two factor with replication ANOVA confirmed difference in viscosities between 0.5 μm PEGylated and carboxylated beads is not statistically significant, with p-value of 0.753. Data are presented as mean values +/− SD, n = 4 videos from one experiment. (e) Viscosity of GRGDSPYS (WT), GQGNSPYS, and GRGNSPYS determined by particle tracking microrheology of 0.5 μm vs. 1 μm bead diameters. Two factor with replication ANOVA confirmed difference in viscosities between 0.5 μm and 1 μm beads is not statistically significant, with p-value of 0.268. Data are presented as mean values +/− SD, n = 4 videos from one experiment.
Extended Data Fig. 6 Sequence based predictors of LLPS.
(a) Solvation free energy from Wolfenden et al.1 vs. saturation concentrations measured in this work. Each data point represents a unique variant used in this work. Variants differing by only one residue are connected by lines such that each mutation results in increasing saturation concentration. Dashed lines indicate variants that follow the trend of preferred interaction with solvent leading to lower phase separation propensity, while solid lines show mutations that result in less favorable interaction with solvent and lower phase separation propensity. (b) Ratio of phase separation propensity score for each sequence relative to the propensity score for WT, calculated using several online sequence-based predictors – DeePhase, PScore, PSpredictor, FuzDrop, LLPhysScore and catgranule. Experimental values are shown as black circles. All predictor values are normalized with the WT to account for different scales used by the predictors. In all cases, when the normalized score is above 1, the sequence is predicted to undergo LLPS more avidly than the WT, while values below 1 indicate a lower propensity to undergo LLPS when compared to WT. Experimental values are calculated from the saturation concentration values (Csat) measured at 37 °C. The experimental values are represented as Csat of WT divided by Csat of variant, such that here too, a value above 1 indicates greater phase separation propensity compared to WT, whereas a value below 1 indicates lower phase separation propensity. (c) Correlation between experimental values and predictor results. Data for all data sets are normalized from 0 to 1. Symbols are the same as shown in (b) for the predictors.
Extended Data Fig. 7 Temperature dependence of saturation concentrations for A-IDP variants.
(a) Ratio of saturation concentrations (Csat) for different sequences with respect to Csat of WT at different temperatures. Lines sloping down indicate that phase separation propensity with respect to the WT is enhanced at higher temperatures, whereas lines sloping upwards indicate reduction in phase separation propensity with respect to WT at higher temperatures. Solid lines indicate the temperatures at which saturation concentration was estimated using turbidimetry experiments, while dashed lines indicate the temperatures at which values were extrapolated from a logarithmic fit to the experimental binodal data. (b) Thermodynamic analysis performed for the different variants based on the estimated saturation concentrations at 20 °C. Higher values indicate greater reduction in phase separation propensity upon carrying out the mutation. Direct estimate refers to values that can be calculated from the experimental variants directly, whereas indirect estimate refers to values for which the mutation was not carried out in this work and the values were inferred based on data from multiple related experimental variants.
Extended Data Fig. 8 Droplet morphology and dynamics after 24 hrs.
(a) Microscopy images for different mutants after 24 hrs of phase separation, showing regular spherical droplets and no signs of fibrillization or aggregation. (Scale bar: 5 µm). Data presented is representative of multiple images acquired for each sample and validated through imaging a second independent sample for 7 out of 12 variants. (b) Microscopy images of GRGNSPYS (cationic sequence), GRGDSPYS (WT) and GQGNSPYS (neutral sequence) undergoing droplet fusion and relaxing into a single spherical droplet, showing liquid-like behavior at 24 hrs. (Scale bar: 2 μm). Data presented is representative of results from two independent trials. Representative WT snapshots from Supplementary Movie 2. (c) Ensemble mean-squared displacement versus lag time at 24 hrs for the three representative variants (GRGNSPYS, GRGDSPYS, and GQGNSPYS), showing liquid-like behavior even after 24 hrs of phase separation.
Supplementary information
Supplementary Information
Supplementary methods, Figs. 1–20, Tables 1–4, references.
Supplementary Video 1
Representative particle tracking microrheology video for WT showing Brownian motion of the 0.5-μm carboxylated fluorescent tracer beads embedded in a droplet. Scale bar, 5 μm.
Supplementary Video 2
Sample video of two WT droplets fusing. Video was recorded after 24 h of phase separation. Video shows droplets contacting, fusing and relaxing into a single spherical droplet even after 24 h of phase separation. Scale bar, 5 μm.
Supplementary Data 1
DNA and peptide sequences for all constructs in this work.
Source data
Source Data Fig. 1
Source data for turbidity plots and phase diagrams for panel b, c and g.
Source Data Fig. 2
Source data for density profile, and contacts for panels b, c and d.
Source Data Fig. 3
Source data for phase diagrams, Csat measurements, contacts of variants for panels a, c, d, e, f and h.
Source Data Fig. 4
Source data for contacts and phase diagrams of variants for panels a, b and d
Source Data Fig. 5
Source data for MSD vs.lag time, viscosity over 8 trials, saturation concentrations and viscosity at 18 °C for panels b, c and d.
Source Data Fig. 6
Source data for saturation concentration values for variants at 37 °C for panel b.
Source Data Extended Data Fig. 1
Source data for salt dependent turbidity and transition temperatures of polycationic variant.
Source Data Extended Data Fig. 2
Source data for pairwise contacts of GQGDSPYS and GRGNSPYS variants.
Source Data Extended Data Fig. 3
Source data for turbidity and partial phase diagrams for GRGASPYA variant.
Source Data Extended Data Fig. 4
Source data for pairwise contacts of GRGNSPWS and GRGNSPFS variants.
Source Data Extended Data Fig. 5
Source data for FRAP, concentration dependent viscosity, MSD and viscosities for surface modified beads and bead size dependence of viscosity measures.
Source Data Extended Data Fig. 6
Source data for A-IDP variant scores from sequence based LLPS predictors.
Source Data Extended Data Fig. 7
Source data for temperature vs. saturation concentration of all variants normalized by WT. Csat changes upon mutation calculated at 20 °C.
Source Data Extended Data Fig. 8
Source data for MSD of three representative sequences at 24-h time point.
Source Data Extended Data Table 1
Source data for saturation concentrations of all variants at 37 °C.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Rekhi, S., Garcia, C.G., Barai, M. et al. Expanding the molecular language of protein liquid–liquid phase separation. Nat. Chem. (2024). https://doi.org/10.1038/s41557-024-01489-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41557-024-01489-x