Abstract
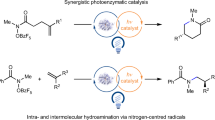
Strategies for achieving asymmetric catalysis with azaarenes have traditionally fallen short of accomplishing remote stereocontrol, which would greatly enhance accessibility to distinct azaarenes with remote chiral centres. The primary obstacle to achieving superior enantioselectivity for remote stereocontrol has been the inherent rigidity of the azaarene ring structure. Here we introduce an ene-reductase system capable of modulating the enantioselectivity of remote carbon-centred radicals on azaarenes through a mechanism of chiral hydrogen atom transfer. This photoenzymatic process effectively directs prochiral radical centres located more than six chemical bonds, or over 6 Å, from the nitrogen atom in azaarenes, thereby enabling the production of a broad array of azaarenes possessing a remote γ-stereocentre. Results from our integrated computational and experimental investigations underscore that the hydrogen bonding and steric effects of key amino acid residues are important for achieving such high stereoselectivities.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Data relating to the materials and methods, optimization studies, experimental procedures, calculations, atomic coordinates, high-performance liquid chromatography spectra and nuclear magnetic resonance spectra are available in Supplementary Information and supplementary data. The structure of OYE1 used for our calculations is available in the PDB database (3TX9). All data are available from the corresponding author upon reasonable request.
References
Carreira, E. M. & Kvaerno, L. Classics in Stereoselective Synthesis (Wiley-VCH, 2008).
Bornscheuer, U. T. et al. Engineering the third wave of biocatalysis. Nature 485, 185–194 (2012).
Devine, P. N. et al. Extending the application of biocatalysis to meet the challenges of drug development. Nat. Rev. Chem. 2, 409–421 (2018).
Chen, K. & Arnold, F. H. Engineering new catalytic activities in enzymes. Nat. Catal. 3, 203–213 (2020).
McGrath, N. A., Brichacek, M. & Njardarson, J. T. A graphical journey of innovative organic architectures that have improved our lives. J. Chem. Educ. 87, 1348–1349 (2010).
Taylor, R. D., MacCoss, M. & Lawson, A. D. G. Rings in drugs. J. Med. Chem. 57, 5845–5859 (2014).
Best, D. & Lam, H. W. C=N-containing azaarenes as activating groups in enantioselective catalysis. J. Org. Chem. 79, 831–845 (2014).
Marcos, V. & Alemán, J. Old tricks, new dogs: organocatalytic dienamine activation of α,β-unsaturated aldehydes. Chem. Soc. Rev. 45, 6812–6832 (2016).
Chauhan, P., Kaya, U. & Enders, D. Advances in organocatalytic 1,6-addition reactions: enantioselective construction of remote stereogenic centers. Adv. Synth. Catal. 359, 888–912 (2017).
Yin, Y., Zhao, X., Qiao, B. & Jiang, Z. Cooperative photoredox and chiral hydrogen-bonding catalysis. Org. Chem. Front. 7, 1283–1296 (2020).
Chai, X. et al. Asymmetric hydroaminoalkylation of alkenylazaarenes via cooperative photoredox and chiral hydrogen-bonding catalysis. Angew. Chem. Int. Ed. 61, e202115110 (2022).
Sorigué, D. et al. An algal photoenzyme converts fatty acids to hydrocarbons. Science 357, 903–907 (2017).
Litman, Z. C., Wang, Y., Zhao, H. & Hartwig, J. F. Cooperative asymmetric reactions combining photocatalysis and enzymatic catalysis. Nature 560, 355–359 (2018).
Liu, X. et al. A genetically encoded photosensitizer protein facilitates the rational design of a miniature photocatalytic CO2-reducing enzyme. Nat. Chem. 10, 1201–1206 (2018).
Sandoval, B. A. & Hyster, T. K. Emerging strategies for expanding the toolbox of enzymes in biocatalysis. Curr. Opin. Chem. Biol. 55, 45–51 (2020).
Grosheva, D. & Hyster, T. K. in Flavin‐Based Catalysis: Principles and Applications (eds Cibulka, R. & Fraaije, M.) 291–313 (Wiley-VCH, 2021).
Harrison, W., Huang, X. & Zhao, H. Photobiocatalysis for abiological transformations. Acc. Chem. Res. 55, 1087–1096 (2022).
Millet, A. et al. Bioinspired supercharging of photoredox catalysis for applications in energy and chemical manufacturing. Acc. Chem. Res. 55, 1423–1434 (2022).
Trimble, J. S. et al. A designed photoenzyme for enantioselective [2+2] cycloadditions. Nature 611, 709–714 (2022).
Sun, N. et al. Enantioselective [2+2]-cycloadditions with triplet photoenzymes. Nature 611, 715–720 (2022).
Emmanuel, M. A., Greenberg, N. R., Oblinsky, D. G. & Hyster, T. K. Accessing non-natural reactivity by irradiating nicotinamide-dependent enzymes with light. Nature 540, 414–417 (2016).
Sandoval, B. A., Meichan, A. J. & Hyster, T. K. Enantioselective hydrogen atom transfer: discovery of catalytic promiscuity in flavin-dependent ‘ene’-reductases. J. Am. Chem. Soc. 139, 11313–11316 (2017).
Toogood, H. S. & Scrutton, N. S. Discovery, characterization, engineering, and applications of ene-reductases for industrial biocatalysis. ACS Catal. 8, 3532–3549 (2018).
Biegasiewicz, K. F. et al. Photoexcitation of flavoenzymes enables a stereoselective radical cyclization. Science 364, 1166–1169 (2019).
Huang, X. et al. Photoenzymatic enantioselective intermolecular radical hydroalkylation. Nature 584, 69–74 (2020).
Page, C. G. et al. Quaternary charge-transfer complex enables photoenzymatic intermolecular hydroalkylation of olefins. J. Am. Chem. Soc. 143, 97–102 (2021).
Huang, X. et al. Photoinduced chemomimetic biocatalysis for enantioselective intermolecular radical conjugate addition. Nat. Catal. 5, 586–593 (2022).
Fu, H. et al. An asymmetric sp3–sp3 cross-electrophile coupling using ‘ene’-reductases. Nature 610, 302–307 (2022).
Kumar Roy, T., Sreedharan, R., Ghosh, P., Gandhi, T. & Maiti, D. Ene-reductase: a multifaceted biocatalyst in organic synthesis. Chem. Eur. J. 28, e202103949 (2022).
Ye, Y. et al. Using enzymes to tame nitrogen-centred radicals for enantioselective hydroamination. Nat. Chem. 15, 206–212 (2023).
Duan, X. et al. A photoenzymatic strategy for radical-mediated stereoselective hydroalkylation with diazo compounds. Angew. Chem. Int. Ed. 62, e202214135 (2023).
Emmanuel, M. A. et al. Photobiocatalytic strategies for organic synthesis. Chem. Rev. 123, 5459–5520 (2023).
Zhang, Z. et al. Photoenzymatic enantioselective intermolecular radical hydroamination. Nat. Catal. 6, 687–694 (2023).
Nakano, Y. et al. Photoenzymatic hydrogenation of heteroaromatic olefins using ‘ene’-reductases with photoredox catalysts. Angew. Chem. Int. Ed. 59, 10484–10488 (2020).
Maestre-Reyna, M. et al. Serial crystallography captures dynamic control of sequential electron and proton transfer events in a flavoenzyme. Nat. Chem. 14, 677–685 (2022).
Himo, F. Recent trends in quantum chemical modeling of enzymatic reactions. J. Am. Chem. Soc. 139, 6780–6786 (2017).
Sheng, X., Kazemi, M., Planas, F. & Himo, F. Modeling enzymatic enantioselectivity using quantum chemical methodology. ACS Catal. 10, 6430–6449 (2020).
Himo, F. & de Visser, S. P. Status report on the quantum chemical cluster approach for modeling enzyme reactions. Commun. Chem. 5, 29 (2022).
Trott, O. A. & Olson, J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461 (2010).
Humphrey, W., Dalke, A. & Schulten, K. VMD: visual molecular dynamics. J. Mol. Graphics 14, 33–38 (1996).
Lu, T. & Chen, F. Multiwfn: a multifunctional wavefunction analyzer. J. Comput. Chem. 33, 580–592 (2012).
Falivene, L. et al. Towards the online computer-aided design of catalytic pockets. Nat. Chem. 11, 872–879 (2019).
Wang, W. & Malcolm, B. A. in In Vitro Mutagenesis Protocols (ed. Braman, J.) 37–43 (Springer, 2002).
Søndergaard, C. R., Olsson, M. H. M., Rostkowski, M. & Jensen, J. H. Improved treatment of ligands and coupling effects in empirical calculation and rationalization of pKa values. J. Chem. Theory Comput. 7, 2284–2295 (2011).
Gaussian 16, Rev. C.02 (Gaussian, 2016).
Becke, A. D. A new mixing of Hartree–Fock and local density functional theories. J. Chem. Phys. 98, 1372–1377 (1993).
Becke, A. D. Density‐functional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 98, 5648–5652 (1993).
Grimme, S., Ehrlich, S. & Goerigk, L. Effect of the damping function in dispersion corrected density functional theory. J. Comput. Chem. 32, 1456–1465 (2011).
Weigend, F. & Ahlrichs, R. Balanced basis sets of split valence, triple zeta valence and quadruple zeta valence quality for H to Rn: design and assessment of accuracy. Phys. Chem. Chem. Phys. 7, 3297–3305 (2005).
Weigend, F. Accurate coulomb-fitting basis sets for H to Rn. Phys. Chem. Chem. Phys. 8, 1057–1065 (2006).
Zhao, Y. & Truhlar, D. G. The M06 suite of density functionals for main group thermochemistry, thermochemical kinetics, non-covalent interactions, excited states, and transition elements: two new functionals and systematic testing of four M06-class functionals and 12 other functionals. Theor. Chem. Acc. 120, 215–241 (2008).
Marenich, A. V., Cramer, C. J. & Truhlar, D. G. Universal solvation model based on solute electron density and on a continuum model of the solvent defined by the bulk dielectric constant and atomic surface tensions. J. Phys. Chem. B 113, 6378–6396 (2009).
Chai, J.-D. & Head-Gordon, M. Long-range corrected hybrid density functionals with damped atom–atom dispersion corrections. Phys. Chem. Chem. Phys. 10, 6615–6620 (2008).
Acknowledgements
This work was funded by the DOE Center for Advanced Bioenergy and Bioproducts Innovation, under the auspices of the US Department of Energy, Office of Science, Office of Biological and Environmental Research (award number DE-SC0018420 to H.Z.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. We acknowledge the Computational Chemistry Commune (http://bbs.keinsci.com/) for their support with DFT calculations. Our research benefitted from the computing resources at Delta, the National Center for Supercomputing Applications, enabled by allocation BIO230016 from the Advanced Cyberinfrastructure Coordination Ecosystem: Services & Support (ACCESS) programme, funded by National Science Foundation grants 2138259, 2138286, 2138307, 2137603 and 2138296. The Delta research computing initiative, funded by the National Science Foundation (award OCI 2005572), the State of Illinois, and a partnership between the University of Illinois at Urbana-Champaign and its National Center for Supercomputing Applications, is deeply appreciated. We are indebted to G. Jiang and H. Cui for their invaluable insights. Special thanks to L. T. Burrus and M. C. O’Dell for their organizational assistance during the project.
Author information
Authors and Affiliations
Contributions
The project was coordinated by H.Z., while H.Z. and M.L. jointly conceptualized the project and designed the research experiments. M.L. executed the experiments and computational studies. W.H. and Z.Z. were responsible for synthesizing some substrates, and Y.Y. contributed to the construction of the mutants.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Chemistry thanks Debabrata Maiti and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Control experiments to evaluate reaction conditions.
Reaction conditions: α-methyl styrene (8 μmol), 2-bromomethyl pyridines (4 μmol), OYE1 (1 mol % based on α-methyl styrene), GDH-105 (5 U/mL), NADP+ (1 mol% based on glucose), glucose (20 μmol), 40 μL DMSO as additives in 1 ml (total) sodium acetate buffer, 445 nm LED, 25 °C, 12 h. The yields were determined by GC-MS. The ee values were determined by HPLC.
Extended Data Fig. 2 Implementation of Quantum Chemical Cluster Approach in OYE1 Protein.
The depicted model excludes the initial substrate. Calculations were executed employing the m062x-D3/def2tzvpp (SMD, ε = 4.0) // b3lyp-D3(BJ)/def2svp (gas) computational level. During the process of geometry optimization, fixed atoms are marked with an asterisk (‘*’). Visual representations were constructed with Pymol (Version 2.5.5) and CLYview 1.0b.
Extended Data Fig. 3 Comprehensive spin density analysis covering intermediates and HAT transition states.
Utilizing Multiwfn 3.8 software, we conducted an in-depth spin density analysis. Visual representations were constructed with VMD (Version 1.9.3). The green and blue areas depict α and β spin respectively, with all amino acid residues (except N194) being omitted to ensure clarity in presentation.
Extended Data Fig. 4 In-depth experimental assessment of steric hindrance in key residues and topographic analysis.
The figure delves into the steric hindrance presented by key residues, particularly focusing on Y375A and T37A. A comprehensive topographic steric map is also provided, with %VBuried indicating the buried volume.
Supplementary information
Supplementary Information
Supplementary Methods, Figs. 1–9, Tables 1–7 and References.
Supplementary Data 1
Computational data for Cartesian coordinates of optimized structures.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, M., Harrison, W., Zhang, Z. et al. Remote stereocontrol with azaarenes via enzymatic hydrogen atom transfer. Nat. Chem. 16, 277–284 (2024). https://doi.org/10.1038/s41557-023-01368-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41557-023-01368-x