Abstract
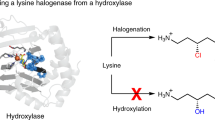
The integration of synthetic and biological catalysis enables new approaches to the synthesis of small molecules by combining the high selectivity of enzymes with the reaction diversity offered by synthetic chemistry. While organohalogens are valued for their bioactivity and utility as synthetic building blocks, only a handful of enzymes that carry out the regioselective halogenation of unactivated \({\rm{C}}_{sp^3}{-}{\rm{H}}\) bonds have previously been identified. In this context, we report the structural characterization of BesD, a recently discovered radical halogenase from the FeII/α-ketogluturate-dependent family that chlorinates the free amino acid lysine. We also identify and characterize additional halogenases that produce mono- and dichlorinated, as well as brominated and azidated, amino acids. The substrate selectivity of this new family of radical halogenases takes advantage of the central role of amino acids in metabolism and enables engineering of biosynthetic pathways to afford a wide variety of compound classes, including heterocycles, diamines, α-keto acids and peptides.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Accession codes for proteins in this study are provided in Supplementary Table 2. The PDB accession code for the BesD structure is 6NIE. Source data are available online for Figs. 3–5. Datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Koeller, K. M. & Wong, C.-H. Enzymes for chemical synthesis. Nature 409, 232–240 (2001).
Kan, S. B. J., Huang, X., Gumulya, Y., Chen, K. & Arnold, F. H. Genetically programmed chiral organoborane synthesis. Nature 552, 132 (2017).
Savile, C. K. et al. Biocatalytic asymmetric synthesis of chiral amines from ketones applied to sitagliptin manufacture. Science 329, 305–309 (2010).
Harris, C., Kannan, R., Kopecka, H. & Harris, T. The role of the chlorine substituent in the antibiotic vancomycin: preparation and characterization of mono- and didechlorovancomycin. J. Am. Chem. Soc. 107, 6652–6658 (1985).
Groll, M., Huber, R. & Potts, B. C. M. Crystal structures of salinosporamide A (NPI-0052) and B (NPI-0047) in complex with the 20S proteasome reveal important consequences of β-lactone ring opening and a mechanism for irreversible binding. J. Am. Chem. Soc. 128, 5136–5141 (2006).
Latham, J., Brandenburger, E., Shepherd, S. A., Menon, B. R. K. & Micklefield, J. Development of halogenase enzymes for use in synthesis. Chem. Rev. 118, 232–269 (2018).
Gkotsi, D. S., Dhaliwal, J., McLachlan, M. M., Mulholand, K. R. & Goss, R. J. Halogenases: powerful tools for biocatalysis (mechanisms applications and scope). Curr. Opin. Chem. Biol. 43, 119–126 (2018).
Neumann, C. S., Fujimori, D. G. & Walsh, C. T. Halogenation strategies in natural product biosynthesis. Chem. Biol. 15, 99–109 (2008).
Marchand, J. A. et al. Discovery of a pathway for terminal-alkyne amino acid biosynthesis. Nature 567, 420–424 (2019).
Vaillancourt, F. H., Yeh, E., Vosburg, D. A., O’Connor, S. E. & Walsh, C. T. Cryptic chlorination by a non-haem iron enzyme during cyclopropyl amino acid biosynthesis. Nature 436, 1191–1194 (2005).
Nakamura, H., Schultz, E. E. & Balskus, E. P. A new strategy for aromatic ring alkylation in cylindrocyclophane biosynthesis. Nat. Chem. Biol. 13, 916–921 (2017).
Anslyn, E. V. & Dougherty, D. A. Modern Physical Organic Chemistry (University Science, 2006).
Agarwal, V. et al. Enzymatic halogenation and dehalogenation reactions: pervasive and mechanistically diverse. Chem. Rev. 117, 5619–5674 (2017).
Liang, T., Neumann, C. N. & Ritter, T. Introduction of fluorine and fluorine-containing functional groups. Ange. Chem. Int. Ed. Engl. 52, 8214–8264 (2013).
Petrone, D. A., Ye, J. & Lautens, M. Modern transition-metal-catalyzed carbon–halogen bond formation. Chem. Rev. 116, 8003–8104 (2016).
Shilov, A. E. & Shul’pin, G. B. Activation of C−H bonds by metal complexes. Chem. Rev. 97, 2879–2932 (1997).
Bollinger, J. M. et al. in 2-Oxoglutarate-Dependent Oxygenases (eds Hausinger R.P. & Schofield, C.J.) 95–122 (Royal Society of Chemistry, London, 2015).
Blasiak, L. C., Vaillancourt, F. H., Walsh, C. T. & Drennan, C. L. Crystal structure of the non-haem iron halogenase SyrB2 in syringomycin biosynthesis. Nature 440, 368–371 (2006).
Mitchell, A. J. et al. Structural basis for halogenation by iron- and 2-oxo-glutarate-dependent enzyme WelO5. Nat. Chem. Biol. 12, 636–640 (2016).
Vaillancourt, F. H., Yin, J. & Walsh, C. T. SyrB2 in syringomycin E biosynthesis is a nonheme FeII α-ketoglutarate- and O2-dependent halogenase. Proc. Natl Acad. Sci. USA 102, 10111–10116 (2005).
Hillwig, M. L. & Liu, X. A new family of iron-dependent halogenases acts on freestanding substrates. Nat. Chem. Biol. 10, 6–10 (2014).
Ortega, M. A. & van der Donk, W. A. New insights into the biosynthetic logic of ribosomally synthesized and post-translationally modified peptide natural products. Cell Chem. Biol. 23, 31–44 (2016).
Runguphan, W., Qu, X. & O’Connor, S. E. Integrating carbon–halogen bond formation into medicinal plant metabolism. Nature 468, 461–464 (2010).
Challis, G. L., Ravel, J. & Townsend, C. A. Predictive, structure-based model of amino acid recognition by nonribosomal peptide synthetase adenylation domains. Chem. Biol. 7, 211–224 (2000).
Dunwell, J. M., Purvis, A. & Khuri, S. Cupins: the most functionally diverse protein superfamily? Phytochemistry 65, 7–17 (2004).
Pandurangan, A. P., Stahlhacke, J., Oates, M. E., Smithers, B. & Gough, J. The SUPERFAMILY 2.0 database: a significant proteome update and a new webserver. Nucleic Acids Res. 47, D490–D494 (2019).
Kulik, H. J. & Drennan, C. L. Substrate placement influences reactivity in non-heme Fe(II) halogenases and hydroxylases. J. Biol. Chem. 288, 11233–11241 (2013).
Matthews, M. L. et al. Substrate-triggered formation and remarkable stability of the C−H bond-cleaving chloroferryl intermediate in the aliphatic halogenase, SyrB2. Biochemistry 48, 4331–4343 (2009).
Puri, M., Biswas, A. N., Fan, R., Guo, Y. & Que, L. Modeling non-heme iron halogenases: high-spin oxoiron(IV)–halide complexes that halogenate C–H bonds. J. Am. Chem. Soc. 138, 2484–2487 (2016).
Galonić, D. P., Barr, E. W., Walsh, C. T., Bollinger, J. M. & Krebs, C. Two interconverting Fe(IV) intermediates in aliphatic chlorination by the halogenase CytC3. Nat. Chem. Biol. 3, 113–116 (2007).
Wong, S. D. et al. Elucidation of the Fe(IV)=O intermediate in the catalytic cycle of the halogenase SyrB2. Nature 499, 320–323 (2013).
Srnec, M. & Solomon, E. I. Frontier molecular orbital contributions to chlorination versus hydroxylation selectivity in the non-heme iron halogenase SyrB2. J. Am. Chem. Soc. 139, 2396–2407 (2017).
Matthews, M. L. et al. Substrate positioning controls the partition between halogenation and hydroxylation in the aliphatic halogenase, SyrB2. Proc. Natl Acad. Sci. USA 106, 17723–17728 (2009).
Mitchell, A. J. et al. Structure-guided reprogramming of a hydroxylase to halogenate its small molecule substrate. Biochemistry 56, 441–444 (2017).
Zhang, Z. et al. Crystal structure of a clavaminate synthase–Fe(II)–2-oxoglutarate–substrate–NO complex: evidence for metal centered rearrangements. FEBS Lett. 517, 7–12 (2002).
Martinie, R. J. et al. Experimental correlation of substrate position with reaction outcome in the aliphatic halogenase, SyrB2. J. Am. Chem. Soc. 137, 6912–6919 (2015).
Gerlt, J. A. Genomic enzymology: web tools for leveraging protein family sequence-function space and genome context to discover novel functions. Biochemistry 56, 4293–4308 (2017).
Matthews, M. L. et al. Direct nitration and azidation of aliphatic carbons by an iron-dependent halogenase. Nat. Chem. Biol. 10, 209–215 (2014).
Fu, G. C. Transition-metal catalysis of nucleophilic substitution reactions: a radical alternative to SN1 and SN2 processes. ACS Cent. Sci. 3, 692–700 (2017).
Nyffeler, P. T., Liang, C.-H., Koeller, K. M. & Wong, C.-H. The chemistry of amine–azide interconversion: catalytic diazotransfer and regioselective azide reduction. J. Am. Chem. Soc. 124, 10773–10778 (2002).
Sletten, E. M. & Bertozzi, C. R. Bioorthogonal chemistry: fishing for selectivity in a sea of functionality. Angew. Chem. Int. Ed. Engl. 48, 6974–6998 (2009).
Roughley, S. D. & Jordan, A. M. The medicinal chemist’s toolbox: an analysis of reactions used in the pursuit of drug candidates. J. Med. Chem. 54, 3451–3479 (2011).
Gatto, G. J., Boyne, M. T., Kelleher, N. L. & Walsh, C. T. Biosynthesis of pipecolic acid by RapL, a lysine cyclodeaminase encoded in the rapamycin gene cluster. J. Am. Chem. Soc. 128, 3838–3847 (2006).
Goodman, J. L. et al. Ornithine cyclodeaminase: structure, mechanism of action, and implications for the μ-crystallin family. Biochemistry 43, 13883–13891 (2004).
Wendisch, V. F., Mindt, M. & Pérez-García, F. Biotechnological production of mono- and diamines using bacteria: recent progress, applications, and perspectives. Appl. Microbiol. Biotechnol. 102, 3583–3594 (2018).
Takatsuka, Y., Yamaguchi, Y., Ono, M. & Kamio, Y. Gene cloning and molecular characterization of lysine decarboxylase from Selenomonas ruminantium delineate its evolutionary relationship to ornithine decarboxylases from eukaryotes. J. Bacteriol. 182, 6732–6741 (2000).
Rudman, D. & Meister, A. Transamination in Escherichia coli. J. Biol. Chem. 200, 591–604 (1953).
Shimizu, Y. et al. Cell-free translation reconstituted with purified components. Nat. Biotechnol. 19, 751–755 (2001).
Edgar, R. C. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5, 113 (2004).
Larsson, A. AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 30, 3276–3278 (2014).
Huang, Y., Niu, B., Gao, Y., Fu, L. & Li, W. CD-HIT Suite: a web server for clustering and comparing biological sequences. Bioinformatics 26, 680–682 (2010).
Jones, D. T., Taylor, W. R. & Thornton, J. M. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8, 275–282 (1992).
Crooks, G. E., Hon, G., Chandonia, J.-M. & Brenner, S. E. WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190 (2004).
Kabsch, W. XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010).
Evans, P. R. & Murshudov, G. N. How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr. 69, 1204–1214 (2013).
Winn, M. D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011).
Skubák, P. & Pannu, N. S. Automatic protein structure solution from weak X-ray data. Nat. Commun. 4, 2777 (2013).
Cowtan, K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D Biol. Crystallogr. 62, 1002–1011 (2006).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 66, 213–221 (2010).
Acknowledgements
This work received support from the National Science Foundation (CHE-1710588) and the Department of Energy (DOE/LBNL DEAC02-05CH11231, FWP CH030201). M.E.N. acknowledges the support of a National Science Foundation graduate research fellowship. J.L.M. acknowledges the support of a National Institutes of Health NRSA training grant (1 T32 GMO66698). J.A.M. acknowledges the support of a University of California, Berkeley Chancellor’s fellowship, Howard Hughes Medical Institute Gilliam fellowship and National Institutes of Health NRSA training grant (1 T32 GMO66698). X-ray data were collected at the Advanced Light Source Beamline 8.3.1, which is operated by the University of California Office of the President, Multicampus Research Programs and Initiatives (MR-15-328599), the National Institutes of Health (R01 GM124149 and P30 GM124169), Plexxikon and the Integrated Diffraction Analysis Technologies program of the U.S. Department of Energy Office of Biological and Environmental Research. The Advanced Light Source is a national user facility operated by Lawrence Berkeley National Laboratory on behalf of the U.S. Department of Energy under contract number DEAC02-05CH11231, Office of Basic Energy Sciences. The funds for the 900-MHz NMR spectrometer housed in the QB3 Institute in Stanley Hall at University of California, Berkeley were provided by the National Institutes of Health (GM68933). We thank E. C. Wittenborn, J. Holton, C. Gee and G. Meigs for crystallography advice. We also thank J.M. Bollinger and A.K. Boal for helpful discussions.
Author information
Authors and Affiliations
Contributions
M.E.N. carried out protein crystallography, bioinformatics and enzyme characterization experiments. K.H.S. carried out enzyme characterization experiments. J.G.P. performed NMR experiments. J.L.M. contributed to bioinformatics. J.A.M. contributed helpful discussions and contributed to bioinformatics. M.E.N., M.C.Y.C. and K.H.S. planned experiments. M.E.N. and M.C.Y.C. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Tables 1–5, Supplementary Figures 1–23 and Supplementary Note 2
Supplementary Data
Cytoscape
Supplementary Note 1
Synthetic Procedures
Rights and permissions
About this article
Cite this article
Neugebauer, M.E., Sumida, K.H., Pelton, J.G. et al. A family of radical halogenases for the engineering of amino-acid-based products. Nat Chem Biol 15, 1009–1016 (2019). https://doi.org/10.1038/s41589-019-0355-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-019-0355-x
This article is cited by
-
A modular and synthetic biosynthesis platform for de novo production of diverse halogenated tryptophan-derived molecules
Nature Communications (2024)
-
Chemodivergent C(sp3)–H and C(sp2)–H cyanomethylation using engineered carbene transferases
Nature Catalysis (2023)
-
Algorithm-aided engineering of aliphatic halogenase WelO5* for the asymmetric late-stage functionalization of soraphens
Nature Communications (2022)
-
Reaction pathway engineering converts a radical hydroxylase into a halogenase
Nature Chemical Biology (2022)
-
Biocatalysis
Nature Reviews Methods Primers (2021)