Abstract
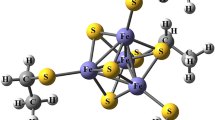
The electronic structure of the nitrogenase metal cofactors is central to nitrogen fixation. However, the P-cluster and FeMo cofactor, each containing eight Fe atoms, have eluded detailed characterization of their electronic properties. We report on the low-energy electronic states of the P-cluster in three oxidation states through exhaustive many-electron wavefunction simulations enabled by new theoretical methods. The energy scales of orbital and spin excitations overlap, yielding a dense spectrum with features that we trace to the underlying atomic states and recouplings. The clusters exist in superpositions of spin configurations with non-classical spin correlations, complicating interpretation of magnetic spectroscopies, whereas the charges are mostly localized from reorganization of the cluster and its surroundings. On oxidation, the opening of the P-cluster substantially increases the density of states, which is intriguing given its proposed role in electron transfer. These results demonstrate that many-electron simulations stand to provide new insights into the electronic structure of the nitrogenase cofactors.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The authors declare that all the data that support the findings of this study are available in the paper and its Supplementary Information, and/or from the authors on reasonable request.
Code availability
The spin-projected and spin-adapted DMRG codes used in this work are available from GitHub repositories (https://github.com/zhendongli2008/zmpo_dmrg and http://sanshar.github.io/Block).
References
Beinert, H., Holm, R. H. & Münck, E. Iron–sulfur clusters: nature’s modular, multipurpose structures. Science 277, 653–659 (1997).
Howard, J. B. & Rees, D. C. Structural basis of biological nitrogen fixation. Chem. Rev. 96, 2965–2982 (1996).
Rees, D. C. & Howard, J. B. The interface between the biological and inorganic worlds: iron–sulfur metalloclusters. Science 300, 929–931 (2003).
Hoffman, B. M., Lukoyanov, D., Yang, Z.-Y., Dean, D. R. & Seefeldt, L. C. Mechanism of nitrogen fixation by nitrogenase: the next stage. Chem. Rev. 114, 4041–4062 (2014).
Spatzal, T. et al. Evidence for interstitial carbon in nitrogenase FeMo cofactor. Science 334, 940–940 (2011).
Lancaster, K. M. et al. X-ray emission spectroscopy evidences a central carbon in the nitrogenase iron–molybdenum cofactor. Science 334, 974–977 (2011).
Sharma, S., Sivalingam, K., Neese, F. & Chan, G. K.-L. Low-energy spectrum of iron–sulfur clusters directly from many-particle quantum mechanics. Nat. Chem. 6, 927–933 (2014).
Chan, J. M., Christiansen, J., Dean, D. R. & Seefeldt, L. C. Spectroscopic evidence for changes in the redox state of the nitrogenase P-cluster during turnover. Biochemistry 38, 5779–5785 (1999).
Danyal, K., Dean, D. R., Hoffman, B. M. & Seefeldt, L. C. Electron transfer within nitrogenase: evidence for a deficit-spending mechanism. Biochemistry 50, 9255–9263 (2011).
Seefeldt, L. C. et al. Energy transduction in nitrogenase. Acc. Chem. Res. 51, 2179–2186 (2018).
Bolin, J. T., Campobasso, N., Muchmore, S. W., Morgan, T. V. & Mortenson, L. E. in Molybdenum Enzymes, Cofactors and Model Systems (eds Stiefel, E., Coucouvanis, D. & Newton, W. E.) Ch. 12, 186–195 (American Chemical Society, 1993).
Peters, J. W. et al. Redox-dependent structural changes in the nitrogenase P-cluster. Biochemistry 36, 1181–1187 (1997).
Keable, S. M. et al. Structural characterization of the P1+ intermediate state of the P-cluster of nitrogenase. J. Biol. Chem. 293, 9629–9635 (2018).
Cao, L., Börner, M. C., Bergmann, J., Caldararu, O. & Ryde, U. Geometry and electronic structure of the P-cluster in nitrogenase studied by combined quantum mechanical and molecular mechanical calculations and quantum refinement. Inorg. Chem. 58, 9672–9690 (2019).
Lanzilotta, W. N., Christiansen, J., Dean, D. R. & Seefeldt, L. C. Evidence for coupled electron and proton transfer in the [8Fe-7S] cluster of nitrogenase. Biochemistry 37, 11376–11384 (1998).
Noodleman, L., Norman, J. G. Jr, Osborne, J. H., Aizman, A. & Case, D. A. Models for ferredoxins: electronic structures of iron-sulfur clusters with one, two, and four iron atoms. J. Am. Chem. Soc. 107, 3418–3426 (1985).
Noodleman, L. & Davidson, E. R. Ligand spin polarization and antiferromagnetic coupling in transition metal dimers. Chem. Phys. 109, 131–143 (1986).
Yamaguchi, K., Fueno, T., Ueyama, N., Nakamura, A. & Ozaki, M. Antiferromagnetic spin couplings between iron ions in iron–sulfur clusters. A localized picture by the spin vector model. Chem. Phys. Lett. 164, 210–216 (1989).
Shoji, M. et al. Theory of chemical bonds in metalloenzymes V: hybrid-DFT studies of the inorganic [8Fe-7S] core. Int. J. Quantum Chem. 106, 3288–3302 (2006).
Schollwöck, U. The density-matrix renormalization group in the age of matrix product states. Ann. Phys. 326, 96–192 (2011).
White, S. R. & Martin, R. L. Ab initio quantum chemistry using the density matrix renormalization group. J. Chem. Phys. 110, 4127–4130 (1999).
Chan, G. K.-L. & Sharma, S. The density matrix renormalization group in quantum chemistry. Annu. Rev. Phys. Chem. 62, 465–481 (2011).
Anderson, P. W. & Hasegawa, H. Considerations on double exchange. Phys. Rev. 100, 675 (1955).
Ohki, Y., Sunada, Y., Honda, M., Katada, M. & Tatsumi, K. Synthesis of the P-cluster inorganic core of nitrogenases. J. Am. Chem. Soc. 125, 4052–4053 (2003).
Coucouvanis, D. et al. Tetrahedral complexes containing the Fe(ii)S4 core. The syntheses, ground-state electronic structures and crystal and molecular structures of the bis(tetraphenylphosphonium)tetrakis (thiophenolato)ferrate(ii) and bis(tetrapheny lphosphonium) bis(dithiosquarato)ferrate(ii) complexes. An analog for the active site in reduced rubredoxins (Rdred). J. Am. Chem. Soc. 103, 3350–3362 (1981).
Noodleman, L. Exchange coupling and resonance delocalization in reduced iron–sulfur [Fe4S4]+ and iron-selenium [Fe4Se4]+ clusters. 1. Basic theory of spin-state energies and EPR and hyperfine properties. Inorg. Chem. 30, 246–256 (1991).
Watt, G. D. & Reddy, K. Formation of an all ferrous Fe4S4 cluster in the iron protein component of Azotobacter vinelandii nitrogenase. J. Inorg. Biochem. 53, 281–294 (1994).
Angove, H. C., Yoo, S. J., Burgess, B. K. & Münck, E. Mössbauer and EPR evidence for an all-ferrous Fe4S4 cluster with S = 4 in the Fe protein of nitrogenase. J. Am. Chem. Soc. 119, 8730–8731 (1997).
Rupnik, K. et al. Nonenzymatic synthesis of the P-cluster in the nitrogenase MoFe protein: evidence of the involvement of all-ferrous [Fe4S4]0 intermediates. Biochemistry 53, 1108–1116 (2014).
Tittsworth, R. C. & Hales, B. J. Detection of EPR signals assigned to the 1-equiv-oxidized P-clusters of the nitrogenase MoFe-protein from Azotobacter vinelandii. J. Am. Chem. Soc. 115, 9763–9767 (1993).
Surerus, K. K. et al. Mössbauer and integer-spin EPR of the oxidized P-clusters of nitrogenase: POX is a non-Kramers system with a nearly degenerate ground doublet. J. Am. Chem. Soc. 114, 8579–8590 (1992).
Owens, C. P., Katz, F. E., Carter, C. H., Oswald, V. F. & Tezcan, F. A. Tyrosine-coordinated P-cluster in G. diazotrophicus nitrogenase: evidence for the importance of O-based ligands in conformationally gated electron transfer. J. Am. Chem. Soc. 138, 10124–10127 (2016).
Shomura, Y., Yoon, K.-S., Nishihara, H. & Higuchi, Y. Structural basis for a [4Fe-3S] cluster in the oxygen-tolerant membrane-bound [NiFe]-hydrogenase. Nature 479, 253 (2011).
Fritsch, J. et al. The crystal structure of an oxygen-tolerant hydrogenase uncovers a novel iron–sulphur centre. Nature 479, 249 (2011).
Volbeda, A. et al. X-ray crystallographic and computational studies of the O2-tolerant [NiFe]-hydrogenase 1 from Escherichia coli. Proc. Natl Acad. Sci. U S A 109, 5305–5310 (2012).
Tabrizi, S. G., Pelmenschikov, V., Noodleman, L. & Kaupp, M. The Mössbauer parameters of the proximal cluster of membrane-bound hydrogenase revisited: a density functional theory study. J. Chem. Theory Comput. 12, 174–187 (2015).
Noodleman, L. & Han, W.-G. Structure, redox, pK a, spin. A golden tetrad for understanding metalloenzyme energetics and reaction pathways. J. Biol. Inorg. Chem. 11, 674–694 (2006).
Lovell, T., Li, J., Liu, T., Case, D. A. & Noodleman, L. FeMo cofactor of nitrogenase: a density functional study of states MN, MOX, MR, and MI. J. Am. Chem. Soc. 123, 12392–12410 (2001).
Dance, I. Electronic dimensions of FeMo-co, the active site of nitrogenase, and its catalytic intermediates. Inorg. Chem. 50, 178–192 (2010).
Siegbahn, P. E. Model calculations suggest that the central carbon in the FeMo-cofactor of nitrogenase becomes protonated in the process of nitrogen fixation. J. Am. Chem. Soc. 138, 10485–10495 (2016).
Bjornsson, R., Neese, F. & DeBeer, S. Revisiting the Mössbauer isomer shifts of the FeMoco cluster of nitrogenase and the cofactor charge. Inorg. Chem. 56, 1470–1477 (2017).
Cao, L. & Ryde, U. Influence of the protein and DFT method on the broken-symmetry and spin states in nitrogenase. Int. J. Quantum Chem. 118, e25627 (2018).
Spatzal, T. et al. Nitrogenase FeMoco investigated by spatially resolved anomalous dispersion refinement. Nat. Commun. 7, 10902 (2016).
Rupnik, K. et al. P+ state of nitrogenase P-cluster exhibits electronic structure of a [Fe4S4]+ cluster. J. Am. Chem. Soc. 134, 13749–13754 (2012).
Mouesca, J.-M., Noodleman, L. & Case, D. Analysis of the 57Fe hyperfine coupling constants and spin states in nitrogenase P-clusters. Inorg. Chem. 33, 4819–4830 (1994).
Mouesca, J.-M., Noodleman, L., Case, D. & Lamotte, B. Spin densities and spin coupling in iron–sulfur clusters: a new analysis of hyperfine coupling constants. Inorg. Chem. 34, 4347–4359 (1995).
Huynh, B. et al. Nitrogenase XII. Mössbauer studies of the MoFe protein from Clostridium pasteurianum W5. Biochim. Biophys. Acta Protein Struct. 623, 124–138 (1980).
Li, Z. & Chan, G. K.-L. Spin-projected matrix product states: versatile tool for strongly correlated systems. J. Chem. Theory Comput. 13, 2681–2695 (2017).
Sharma, S. & Chan, G. K.-L. Spin-adapted density matrix renormalization group algorithms for quantum chemistry. J. Chem. Phys. 136, 124121 (2012).
Acknowledgements
This work was supported by the US National Science Foundation through grant no. NSF-CHE 1665333. The BLOCK and PySCF codes used in the calculations were developed by additional support from the US National Science Foundation through grant no. NSF-SSI 1657286. G.K.-L.C. is a Simons Investigator in Physics.
Author information
Authors and Affiliations
Contributions
Z.L. and G.K.-L.C. designed the study and wrote the manuscript. Z.L. performed the calculations. S.G. contributed to interfacing SP-MPS to SA-MPS used in the BLOCK code. Q.S. contributed to interfacing the PySCF code with COSMO. All authors contributed to the discussion of the results.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Li, Z., Guo, S., Sun, Q. et al. Electronic landscape of the P-cluster of nitrogenase as revealed through many-electron quantum wavefunction simulations. Nat. Chem. 11, 1026–1033 (2019). https://doi.org/10.1038/s41557-019-0337-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41557-019-0337-3
This article is cited by
-
Evaluating the evidence for exponential quantum advantage in ground-state quantum chemistry
Nature Communications (2023)
-
Characterization of charge transfer excited states in [2Fe–2S] iron–sulfur clusters using conventional configuration interaction techniques
Theoretical Chemistry Accounts (2020)