Abstract
Endometrial disorders represent a major gynaecological burden. Current research models fail to recapitulate the nature and heterogeneity of these diseases, thereby hampering scientific and clinical progress. Here we developed long-term expandable organoids from a broad spectrum of endometrial pathologies. Organoids from endometriosis show disease-associated traits and cancer-linked mutations. Endometrial cancer-derived organoids accurately capture cancer subtypes, replicate the mutational landscape of the tumours and display patient-specific drug responses. Organoids were also established from precancerous pathologies encompassing endometrial hyperplasia and Lynch syndrome, and inherited gene mutations were maintained. Endometrial disease organoids reproduced the original lesion when transplanted in vivo. In summary, we developed multiple organoid models that capture endometrial disease diversity and will provide powerful research models and drug screening and discovery tools.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
RNA-seq data were deposited in the Gene Expression Omnibus with accession number GSE118928. Raw sequencing reads of shallow-seq and WES have been deposited in the ArrayExpress database at EMBL−EBI (www.ebi.ac.uk/arrayexpress) under accession numbers E-MTAB-7687 and E-MTAB-7688, respectively. Source data for Fig. 1b–d,g, Fig. 3b–d, Fig. 4b, Fig. 5a,c, Fig. 6a–d, Supplementary Fig. 1d,g-i, Supplementary Fig. 2b, Supplementary Fig. 3c,d, Supplementary Fig. 5d,e,g and Supplementary Fig. 6d−g reported in this study are provided as supplementary source data tables in Supplementary Table 12. All other data supporting the findings of this study are available from the corresponding authors on reasonable request. Unique biological materials can be made available to third parties depending on their research goals (that is, an absence of a conflict of interest), mutual ethical permissions and a Material Transfer Agreement.
References
Roy, A. & Matzuk, M. M. Reproductive tract function and dysfunction in women. Nat. Rev. Endocrinol. 7, 517–525 (2011).
Deligdisch, L. Hormonal pathology of the endometrium. Mod. Pathol. 13, 285–294 (2000).
Vercellini, P., Viganò, P., Somigliana, E. & Fedele, L. Endometriosis: pathogenesis and treatment. Nat. Rev. Endocrinol. 10, 261–275 (2014).
Giudice, L. C. & Kao, L. C. Endometriosis. Lancet 364, 1789–1799.
Siegel, R. L., Miller, K. D. & Jemal, A. Cancer statistics, 2017. CA Cancer J. Clin. 67, 7–30 (2017).
Murali, R., Soslow, R. A. & Weigelt, B. Classification of endometrial carcinoma: more than two types. Lancet Oncol. 15, e268–e278 (2014).
Vollmer, G. Endometrial cancer: experimental models useful for studies on molecular aspects of endometrial cancer and carcinogenesis. Endocr. Relat. Cancer 10, 23–42 (2003).
Depreeuw, J. et al. Characterization of patient-derived tumor xenograft models of endometrial cancer for preclinical evaluation of targeted therapies. Gynecol. Oncol. 139, 118–126 (2015).
Contreras, C. M. et al. Lkb1 inactivation is sufficient to drive endometrial cancers that are aggressive yet highly responsive to mTOR inhibitor monotherapy. Dis. Model. Mech. 3, 181–193 (2010).
King, C. M., Barbara, C., Prentice, A., Brenton, J. D. & Charnock-Jones, D. S. Models of endometriosis and their utility in studying progression to ovarian clear cell carcinoma. J. Pathol. 238, 185–196 (2016).
Boretto, M. et al. Development of organoids from mouse and human endometrium showing endometrial epithelium physiology and long-term expandability. Development 144, 1775–1786 (2017).
Turco, M. Y. et al. Long-term, hormone-responsive organoid cultures of human endometrium in a chemically defined medium. Nat. Cell Biol. 19, 568–577 (2017).
American Society for Reproductive Medicine. Revised American Society for Reproductive Medicine classification of endometriosis: 1996. Fertil. Steril. 67, 817–821 (1997).
Clevers, H. & Nusse, R. Wnt/β-catenin signaling and disease. Cell 149, 1192–1205 (2012).
Pitsos, M. & Kanakas, N. The role of matrix metalloproteinases in the pathogenesis of endometriosis. Reprod. Sci. 16, 717–726 (2009).
Eyster, K. M., Boles, A. L., Brannian, J. D. & Hansen, K. A. DNA microarray analysis of gene expression markers of endometriosis. Fertil. Steril. 77, 38–42 (2002).
Burney, R. O. et al. Gene expression analysis of endometrium reveals progesterone resistance and candidate susceptibility genes in women with endometriosis. Endocrinology 148, 3814–3826 (2007).
Matsuzaki, S. et al. DNA microarray analysis of gene expression profiles in deep endometriosis using laser capture microdissection. Mol. Hum. Reprod. 10, 719–728 (2004).
Weigel, M. T. et al. Differential expression of MMP-2, MMP-9 and PCNA in endometriosis and endometrial carcinoma. Eur. J. Obstet. Gynecol. Reprod. Biol. 160, 74–78 (2012).
Wu, Y. et al. Transcriptional characterizations of differences between eutopic and ectopic endometrium. Endocrinology 147, 232–246 (2006).
Wu, B. et al. Reconstructing lineage hierarchies of mouse uterus epithelial development using single-cell analysis. Stem Cell Rep. 9, 381–396 (2017).
Valentijn, A. J. et al. SSEA-1 isolates human endometrial basal glandular epithelial cells: phenotypic and functional characterization and implications in the pathogenesis of endometriosis. Hum. Reprod. 28, 2695–2708 (2013).
Gonçalves, G. A. et al. p27kip1 overexpression regulates IL-1β in the microenvironment of stem cells and eutopic endometriosis co-cultures. Cytokine 89, 229–234 (2017).
Anglesio, M. S. et al. Cancer-associated mutations in endometriosis without cancer. N. Engl. J. Med. 376, 1835–1848 (2017).
Enomoto, T. et al. Clonal expansion and diversification of cancer-associated mutations in endometriosis and normal endometrium. Cell Rep. 24, 1777–1789 (2018).
Jeong, J.-W. et al. beta-catenin mediates glandular formation and dysregulation of beta-catenin induces hyperplasia formation in the murine uterus. Oncogene 28, 31–40 (2009).
Lynch, H. T., Snyder, C. L., Shaw, T. G., Heinen, C. D. & Hitchins, M. P. Milestones of Lynch syndrome: 1895-2015. Nat. Rev. Cancer 15, 181–194 (2015).
Broutier, L. et al. Human primary liver cancer-derived organoid cultures for disease modeling and drug screening. Nat. Med. 23, 1424–1435 (2017).
Gao, D. et al. Organoid cultures derived from patients with advanced prostate cancer. Cell 159, 176–187 (2014).
Sachs, N. et al. A living biobank of breast cancer organoids captures disease heterogeneity. Cell 172, 373–386 (2018).
Fujii, M. et al. Human intestinal organoids maintain self-renewal capacity and cellular diversity in niche-inspired culture condition. Cell Stem Cell 23, 787–793 (2018).
Aizen, D. et al. Proliferative and signaling activities of insulin analogues in endometrial cancer cells. Mol. Cell. Endocrinol. 406, 27–39 (2015).
Yoshida, S. et al. Induction of hepatocyte growth factor in stromal cells by tumor-derived basic fibroblast growth factor enhances growth and invasion of endometrial cancer. J. Clin. Endocrinol. Metab. 87, 2376–2383 (2002).
Risinger, J. I. et al. PTEN mutation in endometrial cancers is associated with favorable clinical and pathologic characteristics. Clin. Cancer Res. 4, 3005–3010 (1998).
Getz, G. et al. Integrated genomic characterization of endometrial carcinoma. Nature 497, 67–73 (2013).
McConechy, M. K. et al. Use of mutation profiles to refine the classification of endometrial carcinomas. J. Pathol. 228, 20–30 (2012).
Yeramian, A. et al. Endometrial carcinoma: molecular alterations involved in tumor development and progression. Oncogene 32, 403–413 (2013).
Salvesen, H. B. et al. Integrated genomic profiling of endometrial carcinoma associates aggressive tumors with indicators of PI3 kinase activation. Proc. Natl Acad. Sci. USA 106, 4834–4839 (2009).
Van De Wetering, M. et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 161, 933–945 (2015).
Hennes, A. et al. Functional expression of the mechanosensitive PIEZO1 channel in primary endometrial epithelial cells and endometrial organoids. Sci. Rep. 9, 1779 (2019).
Persoons, E. et al. Functional expression of TRP ion channels in endometrial stromal cells of endometriosis patients. Int. J. Mol. Sci. 19, 2467 (2018).
Seino, T. et al. Human pancreatic tumor organoids reveal loss of stem cell niche factor dependence during disease progression. Cell Stem Cell 22, 454–467 (2018).
Stzepourginski, I. et al. CD34+ mesenchymal cells are a major component of the intestinal stem cells niche at homeostasis and after injury. Proc. Natl. Acad. Sci. USA 114, E506–E513 (2017).
Öhlund, D. et al. Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J. Exp. Med. 214, 579–596 (2017).
Pauli, C. et al. Personalized in vitro and in vivo cancer models to guide precision medicine. Cancer Discov. 7, 462–477 (2017).
Girda, E., Huang, E. C., Leiserowitz, G. S. & Smith, L. H. The use of endometrial cancer patient-derived organoid culture for drug sensitivity testing is feasible. Int. J. Gynecol. Cancer 27, 1701–1707 (2017).
Drost, J. et al. Use of CRISPR-modified human stem cell organoids to study the origin of mutational signatures in cancer. Science 358, 234–238 (2017).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Van der Auwera, G. A. et al. From FastQ files to high confidence variant calls: the genome analysis toolkit best practices pipeline. Curr. Protoc. Bioinformat. 43, 1–33 (2013).
Albers, C. A. et al. Dindel: accurate indel calls from short-read data. Genome Res. 21, 961–973 (2011).
Wang, K., Li, M. & Hakonarson, H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010).
Zhao, H.et al. Mismatch repair deficiency endows tumors with a unique mutation signature and sensitivity to DNA double-strand breaks. eLife 3, (2014).
Kim, D. et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013).
De Clercq, K. et al. The functional expression of transient receptor potential channels in the mouse endometrium. Hum. Reprod. 32, 615–630 (2017).
Acknowledgements
We thank our other colleagues (H.V. group: Y. Van Goethem; Stem Cell Institute: A. Santo Ramalho Venâncio; D. Costamagna; P. Peetermans) for their valuable input and technical help. We are also grateful to J. Laureys (Department of Clinical and Experimental Medicine, KU Leuven) and K. Eggermont for their expert help in mouse transplantation experiments, and to the patients, staff and nurses at UZ Leuven for providing the clinical samples. We thank the VIB Nucleomics Core and KU Leuven Genomics Core (particularly Á. Calabuig) for their expert assistance in RNA-seq and aCGH analysis. We are also grateful to InfraMouse (VIB−KU Leuven, Hercules type 3 project ZW09-03) for the use of histological instruments and microscopes. Finally, we acknowledge the use of the Electron Microscopy Platform of the Centre for Human Genetics (VIB−KU Leuven). This work was supported by grants from the KU Leuven Research Fund and from the FWO−Flanders (Belgium). A.H., I.V.Z. and B.C. are supported by a PhD Fellowship from the FWO. M.B. is a PhD Fellow supported by a GOA grant from the Research Fund of the KU Leuven. D.T. is a Senior Clinical Investigator of the FWO.
Author information
Authors and Affiliations
Contributions
M.B. designed the concepts and experiments, performed the experiments and the data analysis and interpreted the results. M.B. and H.V. wrote the manuscript. N.M. contributed to the EC-O protocol set-up, drug screenings, gene expression analyses and routine organoid culturing. X.L. provided essential help in the bioinformatic analysis of RNA-seq data. A.H. performed and cointerpreted the functional ion channel analysis. B.Bo. performed the genomic screening. B.Bo. and D.L. cointerpreted the genomic screening. D.L. supervised the genomic screening. B.Bu., L.P., R.H. and B.C. helped to maintain the organoid cultures. B.Bu. and L.P. helped to perform gene expression analyses. R.H. collected patient information and samples and helped to perform the molecular analyses. H.K. performed and cointerpreted the TEM. I.V.Z. recorded the 3D organoid videos. H.B. guided and supervised the targeted sequencing analysis. M.F. and T.D. added conceptual input. M.F. provided culture essentials. H.U. supervised the 3D imaging. K.P.K. supervised the bioinformatic analyses. A.V., C.M., C.T. and D.T. were driving forces in setting up the clinical collaboration to obtain human samples. I.V. was a collaborating surgeon who provided many of the clinical samples. C.M. and C.T. performed the laparoscopy. C.M., C.T. and D.T. are collaborating gynaecologists with joint grant applications in endometrial research. J.V. supervised and cointerpreted the functional ion channel analysis and is a collaborating scientist with joint grant applications in endometrial research. D.T. performed surgery. H.V. designed and supervised the project, codeveloped the concepts and ideas, codesigned the experiments and coanalysed and cointerpreted the data. All coauthors critically read and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
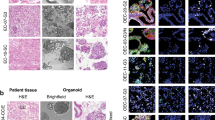
Supplementary Figure 1 Long-term expandable organoids can be established from endometriosis at all stages.
(a) Flowchart depicting organoid establishment from the indicated spectrum of endometrial diseases, downstream analyses and biobanking. (b) Brightfield pictures of development of EM-O and matched ECT-O and EUT-O after seeding (P0). Representative pictures of 3 independent experiments (that is 3 independent donors per condition) are shown. Scale bar, 200 µm. (c) Representative brightfield pictures showing clonal ECT-O formation. Arrow indicates a single cell monitored for 20 days (n=3 biologically independent experiments). Boxed area is magnified. Scale bars, 50 µm. (d) Passaging time of EM-O (3 independent donors) and of matched EUT-O/ECT-O (3 other independent donors), as monitored for 6 passages. Box plot depicts mean, minimum and maximum per individual organoid line. (e) Brightfield and H&E images of organoids derived from rASRM stage I to IV endometriosis. Representative pictures of 3 independent donors per rASRM stage are shown. Scale bars, 200 µm for brightfield and 50 µm for H&E. (f) aCGH plots of short-term (1–2 months; low passage) and long-term (4–6 months; high passage) cultured EM-O and EUT-O. Representative plots of 3 independent experiments are shown. (g) Gene expression analysis of endometrial markers in ECT-O and EM-O after short- and long-term culture, presented as ΔCt (Ct of gene – Ct of GAPDH) (mean ± s.e.m. of n=3 biologically independent experiments). (h) Rescue of IWP2-induced organoid growth inhibition represented as percentage of organoids formed after 10 days with the indicated treatment as compared to SOM (left graphs), and gene expression analysis of canonical (middle) and non-canonical (right) WNT target genes in ECT-O (top) (mean ± s.e.m of n=5 biologically independent experiments) and EM-O (bottom) (mean ± s.e.m of n=4 biologically independent experiments). *P<0.05, **P<0.01, P***<0.001; non-parametric Kruskal-Wallis test for multiple comparison with Dunn’s post-test (95% confidence intervals). (i) WNT ligand gene expression in ECT-O as determined by RT-qPCR and represented as ∆Ct (Ct of gene – Ct of GAPDH) (mean ± s.e.m of 8 independent donors) (left) and as extracted from the RNA-seq dataset and presented as heatmap of transcript per million values (right) in which colors range from white (low) to red (high).
Supplementary Figure 2 Endometriotic organoids show disease characteristics.
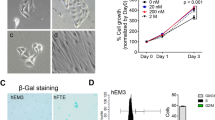
(a) H&E analysis of matched EUT-O and ECT-O. Representative pictures of 5 independent experiments (that is 5 independent donors per condition) are shown. Scale bar, 50 µm. (b) Immunohistochemical analysis of MMP2 and MMP7 in primary peritoneal endometriotic lesions and ECT-O. Representative pictures of 4 independent experiments (that is 4 independent donors per condition) are shown. Scale bar, 50 µm. Gene expression of MMP2 and MMP7, as normalized to GAPDH and expressed as fold change relative to EM-O (mean ± s.e.m. of n=4 biologically independent experiments). *P=0.0286 as compared to EM-O; two-tailed non-parametric T-test (95% confidence intervals). (c) E-cadherin immunofluorescence staining of ECT-O showing the epithelial nature and correct positioning of tight junctions which supports the cells’ polarization. Representative pictures of 5 independent experiments (that is 5 independent donors) are shown. Scale bar, 10 µm. (d) TEM pictures of matched EUT-O and ECT-O from an individual patient. The EUT-O are composed of a single-cell layer bordering a lumen, containing microvilli (magnified box) and ciliated cells (as revealed by acetylated (Ac) α-tubulin immunofluorescence), whereas a stratified, double-cell layer (*) is present in the ECT-O with extensive microvilli (magnified box) and ciliated cells. Representative pictures of 3 independent experiments (that is 3 independent donors per condition) are shown. Scale bars, 10 µm. (e) ECT-O, subrenally transplanted according to the schedule, give rise to lesions with endometriotic features. H&E images and immunohistochemical analysis for ERα and PR of the kidney capsule and ECT-O grafts are presented. Lower panels display H&E staining of the non-grafted kidney, lacking any lesion. Representative pictures of 6 biologically independent experiments with 4 independent donors are shown. Scale bar, 200 µm.
Supplementary Figure 3 Transcriptomic analysis of endometriotic organoids reveals disease-specific pathways and genes.
(a) PCA plot showing the distribution of EM-O, EUT-O and ECT-O, based on the RNA-seq data (n=4 independent donors for EM-O and EUT-O, n=7 independent donors for ECT-O). (b) Diagram of the differentially expressed genes in EM-O, EUT-O and ECT-O as identified by RNA-seq analysis. Of the 277 differentially expressed genes between EM-O/EUT-O and ECT-O, 34 are specifically expressed in EM-O/EUT-O and 243 in ECT-O. Of the 35 differentially expressed genes between EM-O and EUT-O, 15 are specifically expressed in EM-O and 20 in ECT-O (see Supplementary Tables 3–5). Relevant genes are indicated. (c) Expression of WNT ligand and Hippo pathway target genes in stage I to IV ECT-O as normalized to GAPDH and expressed as fold change relative to EM-O (mean ± s.e.m. of n=3 biologically independent experiments for stage I and IV ECT-O, n=5 for stage II ECT-O and n=4 for stage III ECT-O). *P<0.05; non-parametric Kruskal-Wallis test for multiple comparison with Dunn’s post-test (95% confidence intervals). (d) Expression of invasion and inflammatory marker genes in stage I to IV ECT-O as normalized to GAPDH and expressed as fold change relative to EM-O (mean ± s.e.m. of n=3 biologically independent experiments for stage I and IV ECT-O, n=5 for stage II ECT-O and n=4 for stage III ECT-O). *P<0.05; non-parametric Kruskal-Wallis test for multiple comparison with Dunn’s post-test (95% confidence intervals). (e) PCA plot showing the distribution of matched EUT-O and ECT-O, based on the RNA-seq data (n=4 independent donors for the EUT-O and matched ECT-O). (f) Heatmap of 75 differentially expressed genes (log2 normalized counts) as identified by RNA-seq analysis of 4 matched ECT-O and EUT-O organoid lines as indicated. Colors range from blue (low expression) to red (high expression). LGR6 is among the top upregulated genes in the ECT-O.
Supplementary Figure 4 Organoids from endometrial pre-cancer lesions display disease-associated phenotype.
(a) Organoid development from hyperplastic endometrium (HYP-O) after seeding (P0). Representative brightfield pictures of 6 independent experiments (that is 6 independent donors) are shown. Overview (left; scale bar, 200 µm) and magnified organoid pictures (right; scale bar, 50 µm) are presented. (b) H&E analysis, immunohistochemical examination of ERα, PR and P53, and mucin detection (PAS) in primary biopsies and corresponding HYP-O of different types of endometrial hyperplasia as indicated. H&E staining reveals glandular-like morphology with a well-defined lumen in the organoids of simple benign and complex atypical hyperplasia and a poorly-defined lumen in hyperplastic polyp. P53 expression, being present in simple benign hyperplasia and endometrial polyp but absent in complex atypical hyperplasia, is reproduced in the matching organoids. Mucus production is only detected in the lumen of the endometrial polyp and derived organoids (*). Representative brightfield pictures of 2 independent donors for hyperplastic polyp and 3 independent donors for the other hyperplasia types are shown. Scale bars, 50 µm. (c) TEM analysis reveals some stratified epithelium (*). Microvilli are present while cilia are not observed (magnified box). Representative pictures of 3 independent experiments (that is 3 independent donors) are shown. Scale bars, 10 µm. (d) aCGH plots indicate the absence of SCNA in both primary hyperplastic tissue and corresponding HYP-O. Representative plots of 3 independent experiments (that is 3 independent donors) are shown.
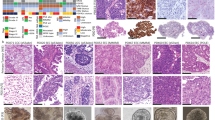
Supplementary Figure 5 EC-derived organoids capture disease and genetic heterogeneity.
(a) EC-O development. Representative pictures of 6 independent experiments (donors). Scale bars, 50 µm. (b) EC-O cultures (P0). Representative pictures of 3 independent donors. Scale bars, 200 µm. (c) aCGH plot of tumor and corresponding organoids, and culture image (n=3 independent donors). (d) Medium optimization. Representative pictures (top) and bar graph (bottom) displaying organoid numbers formed in absence (“-“) of indicated compounds (line and dashed line: SOM for EC-O and HYP-O, respectively) (mean ± s.e.m. of n=3 independent experiments). *P<0.05 among HYP-O as compared to SOM; non-parametric Kruskal-Wallis test with Dunn’s post-test (95% confidence intervals). (e) Organoid passageability (left) (mean ± s.e.m of n=4 independent experiments) and passaging time as monitored for 6 passages (right) (3 independent donors per condition). Box plot depicts mean, minimum and maximum. (f) Representative pictures showing long-term EC-O passaging (n=3 independent donors; magnified organoid in inset). Scale bars, 200 µm (brightfield) and 50 µm (H&E). (g) Clonogenicity (mean organoid numbers for 3 independent donors per group). (h) Different EC-O morphologies (representative pictures of 10 independent cultures). (i) TEM of EC-O showing nuclear abnormalities. Representative pictures of 2 independent experiments. Scale bars, 5 µm. (j) Immunohistochemistry of EC markers and mucin detection (PAS) in primary tumors and corresponding EC-O (each performed in triplicate for 10 independent donors). Scale bars, 50 µm. (k) Flowchart of genomic screen performed on EC-O and primary tumors. (l) aCGH (left) and shallow-seq plot (right), representative of n=8 and n=5 independent donors, respectively. (m) Matrix of SCNA in primary tumor and corresponding organoids. EC-O_12 and EC-O_17 were overtaken by healthy cells (“EC → EM-O”). (n) Genetic aberrations in primary tumors and EC-O. (o) Immunofluorescence of β-catenin in EC-O (n=3 independent donors). Scale bars, 50 µm. Representative pictures of EC-O lines harboring CTNNB1 mutations (EC-O_3), or not (EC-O_6, EC-O_7), cultured in SOM ± XAV939 (n=3 independent donors). Scale bars, 200 µm. (p) Distance matrix based on gene expression data (Supplementary Table 9), from white (identical) to red (divergent). (q) PCA plot based on gene expression analysis (n=4 independent donors for EM-O and HYP-O, n=8 for EC-O, n=4 for primary tumors).
Supplementary Figure 6 EC-derived organoids are amenable to in vivo growth, drug screening, ion channel exploration and biobanking.
(a) Histological analysis of endometrial (cancer) markers in primary EC samples and EC-O-derived xenografts. Representative pictures of n=5 independent donors. Scale bars, 50 µm. (b) Macroscopic pictures of the uterine horns after orthotopic transplantation of samples as indicated (* indicates the non-affected part of the uterine horn). Representative pictures of experiments with 5 independent donors. (c) Immunohistochemical examination of the uterine grafts for the markers indicated. Representative pictures of experiments with 5 independent donors. Scale bars, 50 µm. (d) IC50 of indicated drugs determined from the dose-response curves (Fig. 6a). (e) Gene expression of ion channels in EUT-O and ECT-O subdivided into different rASRM stages, as normalized to the geometric mean of housekeeping genes HPRT1 and PGK1 and expressed as fold change relative to EM-O (mean (± s.e.m. if n≥3) of n=5 EM-O, n=2 EUT-O stage I-II, n=3 EUT-O stage III-IV, n=5 ECT-O stage I-II and n=5 ECT-O stage III-IV independent donors). **P<0.01 compared to EM-O; two-way ANOVA and Dunn’s multiple comparison test. (f) Percentage of responding cells in EM-O, EUT-O and ECT-O to specific ion channel activators. The data represent the following n independent experiments encompassing the indicated total number of cells: GSK in EM-O: n=6, 339 cells; GSK in EUT-O: n=5, 905 cells; GSK in ECT-O: n=4, 458 cells; OAG in EM-O: n=6, 726 cells; OAG in EUT-O: n=4, 772 cells; OAG in ECT-O: n=5, 535 cells; THC in EM-O: n=6, 706 cells; THC in EUT-O: n=4, 959 cells; THC in ECT-O: n=4, 541 cells. (g) Gene expression of ion channels in HYP-O and EC-O, the latter subdivided into non-invasive (N-IN) and invasive (IN) phenotypes, as normalized to HPRT1 and PGK1 and expressed as fold change relative to EM-O (mean ± s.e.m. of n=5 EM-O, n=4 HYP-O, n=3 EC-O N-IN and n=4 EC-O IN independent donors). *P<0.05, **P<0.01, ***P<0.001 compared to EM-O; two-way ANOVA and Dunn’s multiple comparison test. (h) EC-O biobanking with representative organoid line ID card.
Supplementary information
Supplementary Information
Supplementary Figs. 1–6, Supplementary table titles/legends and Supplementary video titles/legends.
Supplementary Table 1
Endometrial organoid biobank.
Supplementary Table 2
SOM for healthy, eutopic, ectopic and hyperplastic endometrium.
Supplementary Table 3
List of genes differentially expressed between EM-O and ECT-O.
Supplementary Table 4
List of genes differentially expressed between EUT-O and ECT-O.
Supplementary Table 5
List of genes differentially expressed between EM-O and EUT-O.
Supplementary Table 6
Top divergent pathways between ECT-O and EM-O as identified by KEGG PATHWAY analysis using the 277 differentially expressed genes.
Supplementary Table 7
Top divergent biological terms between ECT-O and EM-O as identified by DAVID Gene Ontology enrichment using the 277 differentially expressed genes.
Supplementary Table 8
Optimized culture medium for EC-O formation and expansion.
Supplementary Table 9
RT−qPCR gene expression analysis of organoid lines derived from multiple endometrial conditions.
Supplementary Table 10
List of antibodies for immunostaining.
Supplementary Table 11
List of primers for qPCR.
Supplementary Table 12
Statistics source data.
Supplementary Video 1
z-stacks (up to 350 µm) from the bottom to the top of nuclear Hoechst-stained EM-O acquired using two-photon microscopy displaying a single-cell layer surrounding a central lumen.
Supplementary Video 2
z-stacks (up to 350 µm) from the bottom to the top of nuclear Hoechst-stained ECT-O acquired using two-photon microscopy displaying a stratified cell layer surrounding a central lumen.
Supplementary Video 3
z-stacks (up to 350 µm) from the bottom to the top of nuclear Hoechst- and membrane DiI-stained low-grade EC-O acquired using two-photon microscopy displaying a stratified cell layer and the presence of a lumen.
Supplementary Video 4
z-stacks (up to 350 µm) from the bottom to the top of nuclear Hoechst-stained high-grade EC-O combined with brightfield displaying a stratified cell layer and an absence of a lumen.
Rights and permissions
About this article
Cite this article
Boretto, M., Maenhoudt, N., Luo, X. et al. Patient-derived organoids from endometrial disease capture clinical heterogeneity and are amenable to drug screening. Nat Cell Biol 21, 1041–1051 (2019). https://doi.org/10.1038/s41556-019-0360-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-019-0360-z
This article is cited by
-
Patient-derived organoids in human cancer: a platform for fundamental research and precision medicine
Molecular Biomedicine (2024)
-
Integration of pan-omics technologies and three-dimensional in vitro tumor models: an approach toward drug discovery and precision medicine
Molecular Cancer (2024)
-
Transcriptome analysis of adenomyosis eutopic endometrium reveals molecular mechanisms involved in adenomyosis-related implantation failure and pregnancy disorders
Reproductive Biology and Endocrinology (2024)
-
De novo reconstruction of a functional in vivo-like equine endometrium using collagen-based tissue engineering
Scientific Reports (2024)
-
A patient-derived organoid-based study identified an ASO targeting SNORD14E for endometrial cancer through reducing aberrant FOXM1 Expression and β-catenin nuclear accumulation
Journal of Experimental & Clinical Cancer Research (2023)