Abstract
In order to persist, bacteria need to adjust their physiological state in response to external and internal cues. External stimuli are often referred to as stressors. The stringent response, mediated by the alarmone (p)ppGpp, is central to the stress response in many bacteria; yet, there is limited knowledge regarding the role of (p)ppGpp signaling in bacteria belonging to the phylum Bacteroidetes. Like its counterparts in the gut (e.g., Bacteroides thetaiotaomicron and Bacteroides fragilis), Porphyromonas gingivalis persists in close association with its human host. Given the potential for numerous perturbations in the oral cavity, and the fact that P. gingivalis can enter and replicate within host cells, we hypothesized that (p)ppGpp is a key signaling molecule for stress adaptation and persistence. Here, we show that accumulation of ppGpp in P. gingivalis is governed by two homologous enzymes, designated Rel, and RshB, and that ppGpp signaling affects growth rate, survival, biofilm formation, production of outer membrane vesicles, and expression of genes encoding type IX secretion structural and cargo proteins. Overall, our findings provide a potential mechanism by which biofilm formation and virulence of P. gingivalis are integrated via ppGpp signaling, a regulatory mechanism central to bacterial survival in dynamic environments.
Similar content being viewed by others
Introduction
Porphyromonas gingivalis is an anaerobic bacterium belonging to the phylum Bacteroidetes that persists within the oral microbiome, and is strongly associated with the progression of chronic periodontitis.1,2 This bacterium is metabolically unusual, it obtains its iron from heme,3,4 and it does not metabolize sugars; instead, it is highly proteolytic, obtaining its carbon, nitrogen, and energy from protein substrates.5,6,7,8 For this reason, secretion of proteolytic enzymes into its environment is central to P. gingivalis physiology. The type IX secretion system (T9SS), which is restricted to the phylum Bacteroidetes, is a complex translocon that accomplishes the secretion of over 30 P. gingivalis proteins bearing a specific C-terminal domain, including the trypsin-like gingipains, which are key virulence determinants.9 Another key secretion mechanism exploited by P. gingivalis is production of outer membrane vesicles (OMVs). P. gingivalis is highly proficient in OMV production, and not surprisingly, these secreted vesicles are decorated with T9SS cargo proteins, thus providing an effective means to promote the spread of proteolytic enzymes into the surroundings for nutrient acquisition, as well as the spread of virulence determinants.10,11 Although what initiates chronic periodontal disease is still a key question, it is broadly accepted that inflammation and an increased flow of gingival crevicular fluid results in a shift in microbiota to primarily Gram-negative anaerobes, which includes P. gingivalis.12 Recent reports indicate that periodontitis is also strongly linked to systemic inflammatory disorders such as diabetes, Alzheimer’s disease, cardiovascular disease, and rheumatoid arthritis.13,14,15,16,17 In this context, it is important to increase our basic understanding of P. gingivalis physiology in regard to the mechanisms that control its survival and virulence.
Bacteria that persist within the human oral microbiome encounter diverse stressors including pH shifts, oxidative stress, and nutrient limitation, and they adjust their physiology for survival.18,19 The intracellular alarmones, guanosine 5′-diphosphate 3′-diphosphate (ppGpp) and guanosine 5′-triphosphate 3′-diphosphate (pppGpp), are key signaling molecules that enable cell homeostasis.20 It has been reported that changes in the intracellular level of (p)ppGpp can be triggered by various environmental perturbations, including changes in nutrient availability,21 redox,22 pH,23 and temperature.24 The cellular stress response mediated by the increased level of (p)ppGpp is commonly referred to as the stringent response.25 Initial studies on the stringent response focused on the Gram-negative model organism Escherichia coli, which has two multi-domain enzymes: RelA and SpoT.26 RelA-SpoT homologs (RSHs) in other bacteria are key enzymes that synthesize and degrade (p)ppGpp.20,25 The synthase domain (SD) of RSH enzymes catalyzes the transfer of pyrophosphate from ATP to either GDP or GTP to generate ppGpp or pppGpp, respectively, while the RSH hydrolase domain (HD) degrades (p)ppGpp to pyrophosphate and either GDP or GTP. In general, (p)ppGpp levels affect bacterial transcription, translation, and DNA replication, but the mechanisms by which these second messenger nucleotides regulate these cellular processes vary greatly.26,27,28 Studies have also shown that (p)ppGpp signaling is important for diverse biological processes such as antibiotic resistance,29 biofilm formation,30 colonization,31 persistence,32 survival during host invasion,33 and virulence.34,35 Importantly, multiple pathogens use (p)ppGpp signaling to mediate production of virulence factors. The multiplicity of the bacterial genes and regulatory pathways influenced by the (p)ppGpp signaling suggests that the relationship between (p)ppGpp signaling and virulence could be unique for each pathogen. To date, there is limited knowledge regarding the role of (p)ppGpp signaling in bacteria belonging to the phylum Bacteroidetes; in particular, the role of (p)ppGpp signaling in P. gingivalis stress adaptation or virulence has not been investigated. In this study, we generated three deletion mutants in strain 381 in genes predicted to be involved in regulating the intracellular concentrations of ppGpp (Δrel, ΔrshB, and ΔrelΔrshB), and investigated the ppGpp levels and phenotypes in these strains. We compared growth, viability, biofilm formation, morphology, virulence, and the transcriptome between parent strain 381 and its derivatives, and we evaluated the effect of heme deprivation on ppGpp levels. Overall, our data indicate that ppGpp directly or indirectly affects stress adaptation, biofilm matrix composition, production of outer membrane vesicles, and the pathogenic potential of P. gingivalis.
Results
P. gingivalis harbors two enzymes that regulate ppGpp levels whose function is impacted by the availability of hemin
The genome of P. gingivalis strain 381 harbors two genes predicted to encode multi-domain RSH enzymes,18 PGN_0465 (PGF_RS02215 in strain 381) and PGN_1757 (PGF_RS08445 in strain 381). As expected, the genes share greater sequence similarity with BT0700 and BT3998 from Bacteroides thetaiotaomicron, respectively, than they do either to RelA or SpoT from Escherichia coli (Supplementary Fig. 1).31 The data suggest that an RSH gene duplication occurred in bacteria belonging to the phylum Bacteroidetes. Because the nomenclature Rel and RshB for the ancestral bifunctional RSH has been described in the genera Porphyromonas and Bacteroides,20 hereafter we refer to the PGN_0465 and PGN_1757 as Rel and RshB, respectively.
In this study, we generated Δrel, ΔrshB, and ΔrelΔrshB mutants in P. gingivalis strain 381. Interestingly, as shown in Fig. 1a, while no phenotype was observed in the single mutants, the double mutant (ΔrelΔrshB) exhibited less heme-binding capacity (black pigmentation), especially in broth culture. Since heme plays an essential role in the growth, protection against oxidative stress, and virulence of P. gingivalis,3,36 we hypothesized that hemin deprivation could be a key stressor that triggers accumulation of (p)ppGpp in P. gingivalis. To determine whether Rel and RshB are indeed required for (p)ppGpp synthesis, and if hemin has an effect on P. gingivalis (p)ppGpp accumulation, we analyzed 32P-labeled nucleotide extracts by thin-layer chromatography in the presence and absence of 1 µg ml−1 hemin. Since ppGpp and pppGpp accumulate in Enterococcus faecalis in the presence of mupirocin,37 cells of E. faecalis were used as a positive control of ppGpp and pppGpp. As shown in Fig. 1b, P. gingivalis produced ppGpp, which migrates between GTP and the origin; however, unlike E. faecalis, P. gingivalis only produced ppGpp; guanosine pentaphosphate (pppGpp) was not detected. Quantification of ppGpp levels revealed that the parent strain accumulates 2.36 times more ppGpp than the rel mutant and 5.16 times more ppGpp than the rshB mutant. The ppGpp profiles of Δrel and ΔrshB mutants revealed that RshB is the major enzyme responsible for the accumulation of ppGpp, while Rel appears to be responsible for maintaining basal levels under these growth conditions. The nucleotide ppGpp was not detected in the ΔrelΔrshB mutant. Based on these results, we designated the ΔrelΔrshB mutant as ppGpp0.
a When grown overnight to the stationary phase in TSBHK, the ppGpp0 mutant (ΔrelΔrshB) lacks pigmentation, indicative of a defect in heme binding. b Thin-layer chromatography of 32P-labeled extracts from Enterococcus faecalis (control) or P. gingivalis cells in the absence and presence of mupirocin or hemin shows that the ΔrelΔrshB does not synthesize ppGpp, and that the wild type (WT) accumulates ppGpp in response to hemin limitation. Quantification of ppGpp levels was performed by using ImageJ software. The data provided in the lower graph show the average of two biological replicates; error bars depict standard deviation.
Previous attempts by other labs to generate a P. gingivalis ΔrshB mutant were unsuccessful,18,38 and a recent study in B. thetaiotaomicron by Schofield et al. reported that rshB gene (bt3998) was also recalcitrant to transposon insertion, suggesting that rshB-dependent (p)ppGpp hydrolase activity may be required for viability.31 To better understand the probable function of Rel and RshB, we generated a sequence alignment of the Rel and RshB proteins from P. gingivalis and B. thetaiotaomicron along with the (p)ppGpp synthase/hydrolase protein RelSeq from Streptococcus equisimilis (Supplementary Table 1). Since RelSeq has been functionally characterized,39 we used this information to determine if there were any changes in key residues in the hydrolysis and/or synthesis domains. As expected, the Rel and RshB proteins from P. gingivalis and B. thetaiotaomicron showed high identity; yet, the hydrolase domains in Pg_Rel and BT_0700 lacked many of the key residues. Altogether, the data indicate that Rel is a synthase (monofunctional), while RshB has both synthase and hydrolase activity. Importantly, since rshB was recalcitrant to transposon insertion in B. thetaiotaomicron and predicted to be the only RSH with hydrolase activity, we hypothesized that a mutation in rel could have occurred when we generated the rshB mutant in P. gingivalis. To evaluate, we cloned and sequenced rel and its promoter region from the rshB mutant, and verified that the sequence matched the parent strain. In addition, RNA-Seq analysis confirmed that the expression of rel was not downregulated in the rshB mutant. In fact, qPCR analysis (see Fig. 3 below) indicated that rel was slightly upregulated in the rshB mutant, suggesting that under our laboratory growth condition, synthesis of ppGpp by Rel in P. gingivalis strain 381 is low enough to not be inhibitory. Last, our data show that wild-type P. gingivalis increased the level of ppGpp 1.72-fold in response to hemin deprivation, but no ppGpp was detected in the ΔrelΔrshB mutant even under conditions of hemin deprivation (Fig. 1b). Overall, these results show that P. gingivalis strain 381 encodes two RSH proteins, and heme deprivation can activate ppGpp accumulation in P. gingivalis.
Absence of ppGpp resulted in abnormal growth and decreased viability
Recent studies have shown that ppGpp controls a wide range of biological processes such as growth rate and viability.26,40 Having determined that deletion of rel or rshB affects the levels of ppGpp in P. gingivalis, we then evaluated the impact of a lack of ppGpp synthesis and hydrolysis on bacterial growth rate and viability. As shown in Fig. 2a, the ΔrshB and ΔrelΔrshB mutants grew slightly slower than the parent and Δrel strains. Interestingly, in contrast to the other three strains that exhibited a gradual decline during the late stationary phase, the ΔrelΔrshB mutant showed a rapid decline. To verify that this rapid decline in optical density was due to lysis/cell death, we did serial dilutions of the cultures during the growth observation, and spotted 10 μl of dilutions on blood agar plates to determine viable counts (Fig. 2b). As shown in Fig. 2b, the parent strain along with Δrel and ΔrshB showed relatively stable viability from 24 to 120 h, whereas, the ΔrelΔrshB mutant showed a dramatic decrease in viability, as expected.
a P. gingivalis 381 (circles), Δrel (squares), ΔrshB (triangles), and ΔrelΔrshB (crosses) were grown in CDMTHK media. Points indicate the mean values, and error bars indicate standard deviations from three replications. b The ΔrelΔrshB mutant was less viable than other strains in the stationary phase. During the growth observation, cultures of the parent strain 381, Δrel, ΔrshB, and ΔrelΔrshB at 24, 72, and 120 h, were serially diluted, and 10 μl of each dilution from 10−1 to 10−8 was spotted on blood agar plate. The plate was removed from the anaerobic chamber and photographed after 7 days of incubation.
To examine the impact of rel deletion (or complementation) on rshB transcript levels and that of rshB deletion (or complementation) on rel transcript levels, we generated plasmids expressing rel or rshB under the control of their native promoters by using plasmid pT-COW, and transformed them into the corresponding deletion mutants, and determined the expression levels of rel and rshB (qPCR). The relative transcript levels of rel and rshB were determined in the deletion mutants harboring the empty plasmid (pT-COW), pT-rel, or pT-rshB. As shown in Fig. 3, the data indicate that when rel is deleted, rshB is upregulated, and when rshB is deleted, expression of rel is upregulated, when compared with the parent strain (all containing the empty plasmid pT-COW). Further, the data show that when rel or rshB are provided in trans on a plasmid (pT-rel or pT-rshB) in their corresponding mutants, this inversely affects the expression levels of the other gene.
P. gingivalis strain 381 and its derivatives were inoculated in TSBHK and grown for ~24 h to isolate RNA. The relative transcript levels of genes involved in ppGpp accumulation, including Rel (PGN_0465) and RshB (PGN_1757), were determined on the Δrel or ΔrshB deletion mutant harboring the empty plasmid (pT-COW) or pT-rel and pT-rshB. The results are presented as the relative levels (mean ± S.D. of triplicate determinations) compared with the transcript levels of the parent strain 381 harboring the empty plasmid (pT-COW). Error bars represent standard deviations of triplicate replicates.
Since the ΔrelΔrshB double-deletion mutant harbors two antibiotic resistance genes (ermF and tetQ), we generated complementation plasmids that express either rel or rshB under the control of their native promoters using plasmid pC-COW that confers chloramphenicol resistance. We then complemented the ΔrelΔrshB mutant in trans (ΔrelΔrshB/pC-rel or ΔrelΔrshB/pC-rshB). We also generated control strains with the empty vector, pC-COW. Complementation of ΔrelΔrshB partially restored both pigmentation and the growth/viability defects (Fig. 4). Collectively, our studies demonstrate that ppGpp contributes to growth rate and survival of P. gingivalis during the stationary phase.
a When grown overnight to the stationary phase in TSBHK, the ppGpp0 mutant (ΔrelΔrshB) lacks pigmentation, indicative of a defect in heme binding. When the deletion mutant was complemented by any one of the ppGpp synthesis genes from a plasmid (pC-rel or pC-rshB), the pigmentation was partially restored. b P. gingivalis 381/pC-COW (circles), ΔrelΔrshB/pC-COW (crosses), ΔrelΔrshB/pC-rel (squares), and ΔrelΔrshB/pC-rshB (triangles) were grown in CDMTHK media in the presence of chloramphenicol. Points indicate the mean values, and error bars indicate standard deviations from three replications.
Synthesis of ppGpp affects biofilm formation in P. gingivalis
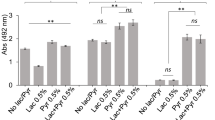
To examine the role of ppGpp in biofilm formation, we compared the biofilm-forming ability of the parent strain in comparison with the mutants in polystyrene microtiter plates. We measured biofilm biomass by safranin staining, and found that the ΔrelΔrshB deletion mutant had an enhanced biofilm phenotype after incubation for 48 h, with an average absorbance at 492 nm (A492) of 0.87 ± 0.02 compared with an A492 of 0.39 ± 0.03 for the parent strain (Fig. 5a). As shown in Fig. 5a, Δrel and ΔrshB mutants showed similar amount of biofilm as the parent strain. Biofilm architectures were compared between the parent strain and ΔrelΔrshB mutant by staining with SYTO 9 to stain all bacterial cells and propidium iodide to detect dead cells. Fluorescence microscopy confirmed that ΔrelΔrshB biofilms were composed of more biomass (Supplementary Fig. 2). The ΔrelΔrshB biofilms were also comprising more dead bacterial cells than parent strain biofilms. The presence of the pC-COW control plasmid lowered the A492 of ΔrelΔrshB to 0.68 ± 0.04 from 0.87 ± 0.02, but ΔrelΔrshB/pC-COW biomass was still significantly greater than that of 381/pC-COW (Fig. 5b). Importantly, complementation of ΔrelΔrshB restored the biofilm to wild-type levels. Taken together, these results show that the absence of ppGpp can enhance biofilm formation in P. gingivalis strain 381.
a The biomass of P. gingivalis 381, Δrel, ΔrshB, and ΔrelΔrshB after 48 h was quantified by staining with safranin. b The biomass of P. gingivalis 381/pC-COW, ΔrelΔrshB/pC-COW, ΔrelΔrshB/pC-rel, and ΔrelΔrshB/pC-rshB after 48 h was quantified by staining with safranin. Data are averages of three replicates (n = 3). Error bars represent the standard deviation. Statistical significance for the ability of biofilm formation was analyzed using the Student’s t test.
The ppGpp0 mutant produces an altered biofilm matrix
Given the results from our previous studies showing that P. gingivalis generates a biofilm with a protein matrix, we hypothesized that the enhanced biofilm phenotype of ΔrelΔrshB strain was due to the accumulation of more matrices. To test this hypothesis, biofilm composition was compared between parent strain and ΔrelΔrshB strain by staining with SYPRO Ruby to detect extracellular protein and SYTO 9 to detect bacterial cells. We found that the biomass of ΔrelΔrshB biofilms was composed of more cells and more protein matrix (Fig. 6). The ratio of extracellular protein to cells (SYPRO Ruby to SYTO 9) was 1.25 ± 0.11 for the parent strain and 2.76 ± 0.40 for ΔrelΔrshB mutant. These results indicate that ΔrelΔrshB biofilms contained more extracellular protein per cell. To further examine the mutants for changes in cell surface properties, we performed transmission electron microscopy (TEM) of negatively stained cells. TEM revealed that ΔrelΔrshB strain produced an abundance of extracellular substance compared with the parent strain (Supplementary Fig. 3a). The surface of the ΔrelΔrshB mutant cells were decorated with negatively stained extracellular substance that extended from the cell surface as fibers to form a meshwork around cells. To investigate the biofilm composition in more detail, we analyzed colony biofilms grown on blood agar plates, using cryo-scanning electron microscopy (Cryo-SEM), as previously described.41 Curiously, in our previous study, we discovered that a peptidylarginine deiminase (PPAD) deletion mutant demonstrated a similar enhanced biofilm phenotype, where the biofilm cells were encased in an extensive meshwork of protein matrix. Using the same growth conditions and methodology, the ΔrelΔrshB colony biofilms showed a remarkably similar biofilm phenotype (Supplementary Fig. 3b).
Parent strain 381 and ΔrelΔrshB mutant were grown for 48 h on glass, and stained with SYTO9 (green color for cells) and SYPRO Ruby (red color for extracellular protein), and three-dimensional z stacks were acquired by fluorescence microscopy. A complete z stack (17 images per stack) was collected using Nikon imaging software (NIS-elements AR 4.30.02, 64-bit). A graticule size: 50 µm (width: 277.41 µm, height: 234.07 µm, and depth: 8 µm). The images shown are representative of three independent experiments. SYTO9, SYPRO Ruby, and total biomass were quantified using Comstat2. Error bars represent the standard deviation. The data were analyzed using the Student’s t test. *p < 0.05.
With the goal of providing more structural information, we added a fixation step to our cryo-SEM protocol. Using this protocol, the colony biofilms of the parent strain showed easily identifiable bacterial cells covered with a large number of outer membrane vesicles (OMVs) (Fig. 7, top). In contrast, the colony biofilms of the ΔrelΔrshB strain showed a smooth surface, and the cells within these biofilms were coated with a matrix-like substance. Importantly, unlike the parent strain, there appear to be very few OMVs produced by the ΔrelΔrshB mutant (Fig. 7, bottom). Overall, the data indicate that the enhanced biofilm phenotype of ΔrelΔrshB strain is due to accumulation of cell surface and extracellular matrix protein(s).
Wild-type strain 381 and ΔrelΔrshB colony biofilms grown anaerobically on blood agar plates for 4 days were imaged by Cryo-SEM. The cells of parent strain 381 were surrounded with a large number of outer membrane vesicles (OMVs); however, the cells of the ΔrelΔrshB strain were coated in an extracellular matrix that lacked OMVs. The middle panels are magnified versions of the left panels. Scale bar: (top left) 5 µm, (top middle) 2 µm, (top right) 2 µm, (bottom left) 5 µm, (bottom middle) 2 µm, and (bottom right) 2 µm.
Virulence of the ppGpp0 mutant is enhanced in the invertebrate Galleria mellonella model
To investigate the significance of ppGpp in P. gingivalis virulence, we used the G. mellonella larvae model that possesses an innate immune system. The Δrel and ΔrshB single mutants killed G. mellonella at rates comparable to the parent strain, with similar averages of larvae survival (~75%) after 78 h of infection (Fig. 8). However, as shown in Fig. 8, virulence of the ΔrelΔrshB mutant was highly enhanced with ~40% larvae survival after 78 h of infection. Consequently, these findings indicate that ppGpp signaling impacts the virulence of P. gingivalis strain 381.
Larvae injected with PBS were used as control. Larvae infected with ΔrelΔrshB mutant showed significantly reduced survival rate compared with the parent strain; *p < 0.05 as determined by Log-rank (Mantel–Cox) test. n = 10 larvae per treatment. The average of two biological replicates is shown; error bars depict standard deviation.
Gene expression is altered in the P. gingivalis ΔrelΔrshB mutant
The gene expression profiles for the parent strain 381 and ΔrelΔrshB mutant were analyzed to identify differentially expressed genes that could account for the various ΔrelΔrshB mutant phenotypes. As shown in Table 1, a total 94 genes were found to be differentially expressed more than 1.5-fold (q value < 0.05) in the ΔrelΔrshB mutant compared with the parent strain. Among these genes, 80 genes were upregulated and 14 genes were downregulated. As expected, the ΔrelΔrshB mutant demonstrated higher expression levels of the transcripts encoding protein synthesis apparatus, including tRNA and ribosomal proteins, a hallmark of a ppGpp0 strain.42 In contrast, the expression of a number of genes involved in heme acquisition was significantly downregulated in the ΔrelΔrshB mutant compared with the parent strain, specifically HagA (PGN_1733) and PGN_1115, which is a predicted hemagglutinin with high sequence identity (46%) to HagA. To further evaluate the relative transcript levels of genes involved in heme acquisition, we performed quantitative PCR (qPCR) analysis on the gene encoding HagA, hemagglutinin PGN_1115, and the gene encoding HagB (PGN_1904). As shown in Supplemental Fig. 4, the data indicate that all three genes are expressed at lower levels in the mutant. In addition, one of the most prominent changes was the expression of genes involved in T9SS-related proteins. Many genes encoding T9SS cargo proteins (PGN_0654, PGN_0657, PGN_0795, PGN_1115, PGN_1733, and PGN_1767) were downregulated. In contrast, genes encoded T9SS structural proteins, including PorV (PGN_0023), PorT (PGN_0778), PorN (PGN_1673), PorM (PGN_1674), PorL (PGN_1675), PorK (PGN_1676), and PorP (PGN_1677), along with PGN_0274—an extracytoplasmic function (ECF) sigma factor (SigP) that regulates the transcriptional level of T9SS genes, and T9SS C-terminal target domain-containing protein (PGN_0852) was upregulated in the ΔrelΔrshB mutant. Notably, the ΔrelΔrshB mutant showed significant upregulation of genes involved in cellular metabolism and biosynthetic process. It has been known that (p)ppGpp is important because of its ability to modify global cellular metabolism nearly instantaneously in response to changes in the external environment, thus optimizing growth and promoting survival.43 Therefore, since ΔrelΔrshB mutant cannot use ppGpp signaling to respond to stress, fitness of ΔrelΔrshB mutant may be impaired due to an imbalance between metabolism and protein secretion. Overall, expression of genes involved in heme acquisition, protein synthesis apparatus, cellular metabolism, and T9SS showed significant changes in the ΔrelΔrshB mutant compared with the parent strain.
Discussion
The natural habitat for P. gingivalis is within the oral microbiome below the gingival margin. This anaerobe not only responds and adapts to many environmental perturbations, such as oxidative stress, but it can also invade host cells and survive within epithelial cells, endothelial cells, and macrophages.44,45 Thus, P. gingivalis has the ability to adapt to, and persist in, a number of habitats. Here, we have explored the impact of a lack of ppGpp synthesis on P. gingivalis physiology and virulence. We have shown that the rel and rshB genes in P. gingivalis encode two RSH proteins that control the levels of ppGpp, and we determined that ppGpp synthesis impacts growth rate, persistence, biofilm formation, and virulence of this strain.
In contrast to the parent strain and the strains with the single mutations, the ΔrelΔrshB double mutant showed a number of specific phenotypes not only for the stringent response with regard to growth and survival, but also for the non-stringent processes, including biofilm formation and virulence. Many studies have documented that (p)ppGpp signaling impacts bacterial survival.46,47,48,49 In this study, only the ΔrelΔrshB mutant showed a decrease in survival during the stationary phase, and this altered phenotype was observed in both basal medium with tryptone (CDMTHK) and in rich TSBHK medium. Since the rel or rshB mutants did not exhibit decreased survival, our results indicate that only basal levels of ppGpp are required for survival of P. gingivalis under prolonged culture conditions; yet, without any ppGpp synthesis P. gingivalis has difficulty in adapting.
Another interesting finding in this study was that P. gingivalis does not appear to accumulate pppGpp. In the previous study by Sen et al., ribosomes were purified from E. coli and P. gingivalis cells, and the (p)ppGpp levels were investigated in these strains using thin-layer chromatography.18 As a positive control, ribosomes isolated from E. coli showed large amounts of both ppGpp and pppGpp. Interestingly, ribosomes isolated from P. gingivalis accumulated significant amount of ppGpp, but no pppGpp was detected as also observed in our study. The previous study mentioned that it is not clear whether the absence of the pppGpp indicates that this molecule is not synthesized in P. gingivalis or that the molecule is unstable. Further investigations are required to determine the reasons for the lack of pppGpp detection in P. gingivalis.
Here, we showed that the putative hemagglutinin PGN_1115, HagA, and HagB were expressed at lower levels in the ΔrelΔrshB mutant when compared with the parent strain (Supplementary Fig. 4). Furthermore, we showed that deletion of rel and rshB resulted in less heme binding (Fig. 1a). This result is consistent with a previous study by Connolly et al., which showed that P. gingivalis W83 exhibited typical black colonies, whereas ΔhagBΔhagC mutant produced white colonies.3 In addition, our results showed that the parent strain increased the level of ppGpp in response to hemin starvation (Fig. 1b). Overall, these results support our working hypothesis that ppGpp signaling directly or indirectly regulates the mechanisms involved in heme acquisition in P. gingivalis.
One of the most remarkable discoveries of this study was that ppGpp impacts biofilm matrix composition. Our data show that the ΔrelΔrshB mutant forms a biofilm with more total biomass (~2-fold), yet the biofilms consisted of a higher proportion of dead cells. Moreover, the increase in biomass was not only from more cells (dead or alive); in addition, the biofilms consisted of more protein matrix. Furthermore, the matrix composition was also distinct. The matrix of the wild-type biofilm contains copious amounts of OMVs, while the OMV content was negligible in the biofilms formed by the mutant, indicating that ppGpp synthesis affects OMV production. Since transcriptomic analysis discovered that expression of genes encoding the type IX secretion structural proteins was significantly increased in the ΔrelΔrshB mutant, it is likely that the enhanced extracellular matrix produced by ΔrelΔrshB mutant is made up of proteins secreted by the T9SS (Fig. 9). T9SS is found only in the Bacteroidetes phylum. This novel secretion system plays a role in gliding motility,50 and secretion of virulence factors,51 and as we recently reported, it is central to P. gingivalis surface translocation.52 In P. gingivalis, T9SS transports select proteins across the outer membrane that are involved in virulence, e.g., gingipains RgpA, RgpB, and Kgp.9,51 As shown in Table 1, RNA-seq analysis revealed that an extracytoplasmic function (ECF) sigma factor (PGN_0274) was significantly upregulated in the ΔrelΔrshB mutant. ECF sigma factors facilitate changes in gene expression by binding to and guiding the core RNA polymerase. The ECF sigma factor SigP (PGN_0274) has been shown to regulate expression of gingipains, biofilm formation, and the T9SS.53 Thus, although many T9SS cargo proteins are downregulated in the ppGpp0 mutant, it is possible that the enhanced virulence is due to overproduction of a T9SS-secreted virulence factor. An alternative hypothesis is that dead cells may release internal components, such as peptidoglycan, which for P. gingivalis has been shown to be quite toxic.54 Additional studies are required to identify the true impact of ppGpp signaling on the T9SS and the pathogenicity of P. gingivalis.
When ppGpp is not synthesized (ppGpp0), the expression of genes involved in heme acquisition is downregulated, while there are higher expression levels of genes involved in protein synthesis (tRNA and ribosomal proteins). The ECF sigma factor SigP, which positively regulates T9SS (por genes), is also significantly upregulated in response to a lack of ppGpp. Our working model is that ppGpp synthesis indirectly regulates secretion and localization proteins secreted via T9SS, which in turn impacts outer membrane vesicle production and biofilm matrix formation.
This study provides new insights into how ppGpp impacts gene expression in P. gingivalis, and increases our understanding of functional links between ppGpp signaling and the virulence of this bacterium. Importantly, our studies suggest that when P. gingivalis is intrinsically stressed, i.e., it is unable to synthesize a stress adaptation signal (ppGpp0 mutant) or it cannot control the levels of arginine (PPAD mutant), this results in enhanced biofilm formation and a decrease in the production of OMVs. Future studies will determine the significance of this phenotype, but it is intriguing to speculate that being able to inversely regulate protein matrix formation and OMV production may be fundamental to the survival of this bacterium.
Methods
Bacterial strains, mutant construction, and complementation
P. gingivalis strain 381 (provided by Dr. Howard Kuramitsu, State University of Buffalo, Buffalo, NY) and derivatives were grown on agar plates containing Trypticase Soy Broth (Becton, Dickinson and Company, Franklin Lakes, NJ, USA) supplemented with 5 µg ml−1 hemin, 1 µg ml−1 menadione, and 5% defibrinated sheep blood (BAPHK) (Northeast Laboratory Services, Winslow, ME, USA) at 37 °C in an anaerobic chamber (Coy Lab Products, Grass Lake, MI, USA) with an atmosphere containing 5% hydrogen, 10% carbon dioxide, and 85% nitrogen. Since the high sequence similarity between P. gingivalis strain ATCC 33277 and strain 381, and the nomenclature of genes in strain ATCC 33277 are more detailed than strain 381, we used the names of genes from the strain ATCC 33277 in this report. Mutant strains 381 Δrel, 381 ΔrshB, and 381 ΔrelΔrshB were generated as previously described.55 Briefly, primers were designed to generate upstream and downstream products of ~1 kb flanking rel or rshB, as well as an erythromycin resistance gene (ermF) obtained from plasmid pVA2198,56 or a tetracycline resistance gene (tetQ) obtained from pT-COW.57,58 All primers used in this study are listed in Supplementary Table 2. These oligonucleotides were used to prime PCRs using genomic DNA from P. gingivalis strain 381 and Phusion high-fidelity PCR master mix with HF buffer according to the manufacturer’s instructions. The products were purified and combined using the NEBuilder HiFi DNA Assembly Master Mix (New England BioLabs, Ipswich, MA, USA) according to the instructions provided by the manufacturer. The final product was mixed with previously frozen cells of P. gingivalis and transformed by electroporation.59 P. gingivalis deletion mutants were maintained by supplementing media with 10 µg ml−1 erythromycin or 1 µg ml−1 tetracycline. Complementation of the ΔrelΔrshB mutant was performed by inserting rel (PGN_0465) or rshB (PGN_1757) under the control of their native promoter regions into plasmid pC-COW (graciously provided by Ana Duran-Pinedo, University of Florida, Gainesville, FL), generating pC-rel and pC-rshB, respectively. Complemented strains were generated by conjugation as previously described.60 In brief, BAPHK containing chloramphenicol (10 µg ml−1) was used to select for pC-COW containing P. gingivalis strains, and gentamicin (200 µg ml−1) was used to counterselect the E. coli S17-1 donor. Transconjugants were obtained after 7 days of anaerobic incubation. Clones were isolated, verified by PCR, and maintained on BAPHK containing chloramphenicol (10 µg ml−1). To complement the rel or rshB single-deletion mutants, rel or rshB were cloned under the control of their native promoter regions into plasmid pT-COW, generating pT-rel and pT-rshB, respectively. Complemented strains were maintained on BAPHK containing tetracycline (1 µg ml−1). Details of bacterial strain and plasmid constructions are provided in Supplementary Table 3.
Bacterial growth
Broth cultures of P. gingivalis were grown anaerobically in Tryptic Soy Broth (TSB) medium (Becton, Dickinson and Company, Franklin Lakes, NJ, USA) supplemented with 5 µg ml−1 hemin and 1 µg ml−1 menadione (TSBHK). Cultures grown overnight in TSBHK were diluted 1:125 in pre-reduced chemically defined medium (CDM) supplemented with 1% tryptone (CDMT), 5 µg ml−1 hemin, and 1 µg ml−1 menadione (CDMTHK). Bacterial growth was then monitored by measuring the optical density at 600 nm, and presented as the mean ± standard deviations (n = 3).
Detection of (p)ppGpp accumulation patterns
For (p)ppGpp measurements in P. gingivalis, overnight cultures grown in TSBHK were diluted 1:125 in pre-reduced CDMTHK. The 0.9-ml cultures of each strain were grown to an optical density at 600 nm of ≈0.5, and prelabeled with 100 μCi of carrier-free [32P]orthophosphate (Amersham Biosciences, Piscataway, NJ) aerobically for 4 h at 37 °C. Samples were centrifuged at 13,000 × g for 1 min, and 880 μl of the supernatants were discarded. Nucleotide pools were extracted by adding 50 μl of 3 M ice-cold formic acid, followed by two freeze–thaw cycles. Acid extracts were centrifuged briefly, and 30 μl of the supernatant fluids were spotted onto polyethyleneimine (PEI)–cellulose plates (Selecto Scientific, Inc., Suwanee, GA) for separation by thin-layer chromatography (TLC) in 1.25 M KH2PO4 (pH 3.4). After 30 min of air-drying, the TLC plate was carefully wrapped and exposed to the X-ray film in the radiation cassette for 72 h. The levels of ppGpp were measured using ImageJ software by determining the pixel intensity in each strain.
Growth and imaging for biofilms
Biofilm assays were performed as previously described with slight modifications.61 Briefly, P. gingivalis cultures were grown anaerobically in Todd Hewitt Broth (THB) medium (Becton, Dickinson and Company, Franklin Lakes, NJ, USA) supplemented with 5 µg ml−1 hemin and 1 µg ml−1 menadione (THBHK) for 24 h at 37 °C, subcultured into pre-reduced CDMTHK, and grown to an OD600 of ≈1.0, diluted in a pre-reduced CDMTHK to OD600 0.2. Culture aliquots of 200 µl were then placed into uncoated 96-well polystyrene flat-bottom plates under anaerobic conditions at 37 °C for 48 h. Fluorescent imaging was performed in the anaerobic chamber on biofilms grown in 16-well removable coverglass (Grace Bio-Labs, Inc. Bend, Oregon, USA) using SYPRO Ruby Biofilm Matrix Stain (Invitrogen by Thermo Fisher Scientific) and the Invitrogen Live/Dead BacLight Bacterial Viability Kit (Invitrogen by Thermo Fisher Scientific) as per the manufacturer’s instructions, as previously described41,62 with slight modification. In brief, culture supernatants (0.2 mL) were removed; wells were washed twice with distilled water; 0.2 mL of mixture (components A [SYTO9] and B [propidium iodide or SYPRO Ruby] were mixed in equal volumes, and 0.6 µl were added per 0.2 ml of distilled water) was added to each well; then the plate was incubated in the dark for 15 min. The dye mixture was removed; wells were washed twice with distilled water; 50 μl of water was added to cover the base of the well. Images of biofilms stained with SYTO9 and propidium iodide, or SYPRO Ruby, were acquired using a Nikon Eclipse Ti inverted fluorescence microscope. SYTO9 fluorescence was detected using the FITC band-pass filter cube. Propidium iodide or SYPRO Ruby fluorescence was detected using the Texas Red bandpass filter cube. Stacked maximum intensity projection images were analyzed using Comstat2 version 2.1.
Cryo-scanning electron microscopy (Cryo-SEM) of biofilms
Electron microscopy and image analysis were performed by the electron microscopy core of Interdisciplinary Center for Biotechnology Research (ICBR) at the University of Florida. Cryo-SEM experiments were performed using a Quorum PP3010T cryotransfer system (Quorum Technologies, Electron Microscopy Sciences) attached to a Hitachi SU5000FE VP-SEM (Hitachi High Technologies, America). Samples were transported to the UF ICBR EM Core under anaerobic conditions using AnaeroPack (Thermo Scientific) sachets inside Kapak SealPAK Pouches (VWR) and heat-sealed. Colonies were immediately fixed by immersion with 4% formaldehyde and 1% glutaraldehyde in 0.1 M cacodylate buffer, pH 7.24, and kept at 4 °C overnight. Samples were prepared for Cryo-SEM by removal of the colony biofilm from blood agar substrate with a small spatula, and mounted on the edge into the 1-mm slot of a copper stub, with a mixture of colloidal graphite and OCT low-temperature adhesive (Electron Microscopy Sciences). After attaching the sample stub to a transfer shuttle, the colony was vitrified in liquid ethane within a liquid nitrogen reservoir. The ethane-frozen sample was then rapidly plunged into the PrepDek® workstation liquid nitrogen slush at −210 °C under vacuum, and immediately transferred to the Cryo-preparation chamber. The side-mounted colony was fractured, sublimed at −60 °C for 15 min, and sputter coated with platinum for 60 s at 10-mA current in an argon atmosphere. The Cryo-prep chamber returned to −140 °C, at a vacuum of >10−5 mbar, and transferred to the nitrogen gas-cooled cold stage inside the SEM chamber. The sample remained frozen during the imaging at −140 °C, under high vacuum conditions using 5-keV, current emission 194,000 nA, and working distance between 5 and 10 mm.
G. mellonella model of systemic infection
Larvae of G. mellonella was used to assess virulence of P. gingivalis 381 and its derivatives as described previously.42 Briefly, groups of 10 larvae (200–300 mg in weight) were injected with 5 μl of bacterial inoculum containing ~3 × 107 CFU. After injection, larvae were kept at 37 °C, and G. mellonella survival was recorded at selected intervals for up to 78 h. The results were analyzed with GraphPad Prism 4.0 software. Experiments were performed independently at least two times with similar results.
RNA extraction, sequencing, and quantitative PCR (qPCR) analysis
P. gingivalis strain 381 and its derivatives were inoculated in TSBHK, and grown for ~24 h. The cultures were subcultured in pre-reduced TSBHK, and grown to an OD600 of 1.0. Cultures were diluted 1/5 with fresh pre-reduced TSBHK, and 10-μl aliquots of each culture were spotted on blood agar plates. After 24 h of incubation, cultures were scraped off the plates, and the RNA extraction was performed using the Direct-zol RNA Miniprep kit (Zymo Research), according to the instructions provided by the manufacturer with a slight modification.63 RNA samples were delivered to the Gene Expression and Genotyping core of ICBR at the University of Florida. Sample quality determination and sequencing were performed by gene expression and genotyping core in the ICBR.52 The program ‘Rockhopper’64 was used for aligning sequencing reads to the genome reference of P. gingivalis 381. We eliminated any of the sequencing reads with a q value > 0.05 and a fold change of <1.5. The qPCR was performed as described previously.65 Briefly, cDNA was produced from the same amount of RNA from each sample by using cDNA EcoDry Premix (Clontech). cDNAs were mixed with gene-specific primers and iQ SYBR Green Supermix (Bio-Rad). The qPCR was performed using the CFX96 Real-Time System (Bio-Rad).
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The authors declare that the data supporting the findings of this study are available within the paper and its Supplementary Information files. Raw sequencing data are available on the NCBI Sequence Read Archive (SRA) under accession number PRJNA600517.
References
Socransky, S. S. & Haffajee, A. D. Periodontal microbial ecology. Periodontol 2000 38, 135–187 (2005).
Socransky, S. S., Haffajee, A. D., Cugini, M. A., Smith, C. & Kent, R. L. Jr. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 25, 134–144 (1998).
Connolly, E. et al. The Porphyromonas gingivalis hemagglutinins HagB and HagC are major mediators of adhesion and biofilm formation. Mol. Oral. Microbiol 32, 35–47 (2017).
Olczak, T., Simpson, W., Liu, X. & Genco, C. A. Iron and heme utilization in Porphyromonas gingivalis. FEMS Microbiol Rev. 29, 119–144 (2005).
Grenier, D., Gauthier, P., Plamondon, P., Nakayama, K. & Mayrand, D. Studies on the aminopeptidase activities of Porphyromonas gingivalis. Oral. Microbiol Immunol. 16, 212–217 (2001).
Lamont, R. J. & Jenkinson, H. F. Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol Mol. Biol. Rev. 62, 1244–1263 (1998).
Oda, H., Saiki, K., Tonosaki, M., Yajima, A. & Konishi, K. Participation of the secreted dipeptidyl and tripeptidyl aminopeptidases in asaccharolytic growth of Porphyromonas gingivalis. J. Periodontal Res. 44, 362–367 (2009).
Vansteenbergen, T. J. M. & Degraaff, J. Proteolytic activity of black-pigmented bacteroides strains. Fems Microbiol. Lett. 33, 219–222 (1986).
Nakayama, K. Porphyromonas gingivalis and related bacteria: from colonial pigmentation to the type IX secretion system and gliding motility. J. Periodontal Res. 50, 1–8 (2015).
Gui, M. J., Dashper, S. G., Slakeski, N., Chen, Y. Y. & Reynolds, E. C. Spheres of influence: Porphyromonas gingivalis outer membrane vesicles. Mol. Oral. Microbiol 31, 365–378 (2016).
Veith, P. D., Luong, C., Tan, K. H., Dashper, S. G. & Reynolds, E. C. Outer membrane vesicle proteome of porphyromonas gingivalis is differentially modulated relative to the outer membrane in response to heme availability. J. Proteome Res. 17, 2377–2389 (2018).
Marsh, P. D. Are dental diseases examples of ecological catastrophes? Microbiology 149, 279–294 (2003).
Singhrao, S. K. & Olsen, I. Assessing the role of Porphyromonas gingivalis in periodontitis to determine a causative relationship with Alzheimer’s disease. J. Oral. Microbiol 11, 1563405 (2019).
Scher, J. U. & Abramson, S. B. Periodontal disease, Porphyromonas gingivalis, and rheumatoid arthritis: what triggers autoimmunity and clinical disease? Arthritis Res. Ther. 15, 122 (2013).
Olsen, I. & Yilmaz, O. Possible role of Porphyromonas gingivalis in orodigestive cancers. J. Oral Microbiol. 11, https://doi.org/10.1080/20002297.2018.1563410 (2019).
Koziel, J., Mydel, P. & Potempa, J. The link between periodontal disease and rheumatoid arthritis: an updated review. Curr. Rheumatol. Rep. 16, 408, https://doi.org/10.1007/s11926-014-0408-9 (2014).
Carter, C. J., France, J., Crean, S. & Singhrao, S. K. The Porphyromonas gingivalis/Host Interactome Shows Enrichment in GWASdb Genes Related to Alzheimer’s Disease, Diabetes and Cardiovascular Diseases. Front Aging Neurosci. 9, 408 (2017).
Sen, K., Hayashi, J. I. & Kuramitsu, H. K. Characterization of the relA gene of Porphyromonas gingivalis. J. Bacteriol. 182, 3302–3304 (2000).
Kim, H. M., Waters, A., Turner, M. E., Rice, K. C. & Ahn, S. J. Regulation of cid and lrg expression by CcpA in Streptococcus mutans. Microbiology 165, 113–123 (2019).
Atkinson, G. C., Tenson, T. & Hauryliuk, V. The RelA/SpoT homolog (RSH) superfamily: distribution and functional evolution of ppGpp synthetases and hydrolases across the tree of life. PLoS ONE 6, e23479 (2011).
Traxler, M. F. et al. Discretely calibrated regulatory loops controlled by ppGpp partition gene induction across the ‘feast to famine’ gradient in Escherichia coli. Mol. Microbiol. 79, 830–845 (2011).
Glass, T. L., Holmes, W. M., Hylemon, P. B. & Stellwag, E. J. Synthesis of guanosine tetra- and pentaphosphates by the obligately anaerobic bacterium Bacteroides thetaiotaomicron in response to molecular oxygen. J. Bacteriol. 137, 956–962 (1979).
Wells, D. H. & Gaynor, E. C. Helicobacter pylori initiates the stringent response upon nutrient and pH downshift. J. Bacteriol. 188, 3726–3729 (2006).
Gallant, J., Palmer, L. & Pao, C. C. Anomalous Synthesis of ppGpp in Growing Cells. Cell 11, 181–185 (1977).
Potrykus, K. & Cashel, M. (p)ppGpp: still magical? Annu Rev. Microbiol 62, 35–51 (2008).
Hauryliuk, V., Atkinson, G. C., Murakami, K. S., Tenson, T. & Gerdes, K. Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat. Rev. Microbiol 13, 298–309 (2015).
Liu, K., Bittner, A. N. & Wang, J. D. Diversity in (p)ppGpp metabolism and effectors. Curr. Opin. Microbiol 24, 72–79 (2015).
Steinchen, W. & Bange, G. The magic dance of the alarmones (p)ppGpp. Mol. Microbiol 101, 531–544 (2016).
Poole, K. Bacterial stress responses as determinants of antimicrobial resistance. J. Antimicrobial Chemother. 67, 2069–2089 (2012).
Liu, H., Xiao, Y., Nie, H., Huang, Q. & Chen, W. Influence of (p)ppGpp on biofilm regulation in Pseudomonas putida KT2440. Microbiol Res. 204, 1–8 (2017).
Schofield, W. B., Zimmermann-Kogadeeva, M., Zimmermann, M., Barry, N. A. & Goodman, A. L. The stringent response determines the ability of a commensal bacterium to survive starvation and to persist in the gut. Cell Host Microbe 24, 120-+ (2018).
Maisonneuve, E. & Gerdes, K. Molecular mechanisms underlying bacterial persisters. Cell 157, 539–548 (2014).
Geiger, T. et al. The stringent response of Staphylococcus aureus and Its Impact on survival after phagocytosis through the induction of intracellular PSMs expression. PLoS Pathog. 8, e1003016 (2012).
Dalebroux, Z. D., Svensson, S. L., Gaynor, E. C. & Swanson, M. S. ppGpp conjures bacterial virulence. Microbiol. Mol. Biol. Rev. 74, 171-+ (2010).
Dozot, M. et al. The stringent response mediator Rsh is required for Brucella melitensis and Brucella suis virulence, and for expression of the type IV secretion system virB. Cell. Microbiol. 8, 1791–1802 (2006).
Anaya-Bergman, C., Rosato, A. & Lewis, J. P. Iron- and hemin-dependent gene expression of Porphyromonas gingivalis. Mol. Oral. Microbiol. 30, 39–61 (2015).
Abranches, J. et al. The molecular alarmone (p)ppGpp mediates stress responses, vancomycin tolerance, and virulence in Enterococcus faecalis. J. Bacteriol. 191, 2248–2256 (2009).
Klein, B. A. et al. Identification of essential genes of the periodontal pathogen Porphyromonas gingivalis. BMC Genomics 13, 578 (2012).
Hogg, T., Mechold, U., Malke, H., Cashel, M. & Hilgenfeld, R. Conformational antagonism between opposing active sites in a bifunctional RelA/SpoT homolog modulates (p)ppGpp metabolism during the stringent response [corrected]. Cell 117, 57–68 (2004).
Li, G. et al. Role of (p)ppGpp in viability and biofilm formation of Actinobacillus pleuropneumoniae S8. PLoS ONE 10, e0141501 (2015).
Vermilyea, D. M., Ottenberg, G. K. & Davey, M. E. Citrullination mediated by PPAD constrains biofilm formation in P. gingivalis strain 381. NPJ Biofilms Microbiomes 5, 7 (2019).
Gaca, A. O., Abranches, J., Kajfasz, J. K. & Lemos, J. A. Global transcriptional analysis of the stringent response in Enterococcus faecalis. Microbiology 158, 1994–2004 (2012).
Srivatsan, A. & Wang, J. D. Control of bacterial transcription, translation and replication by (p)ppGpp. Curr. Opin. Microbiol 11, 100–105 (2008).
Bostanci, N. & Belibasakis, G. N. Porphyromonas gingivalis: an invasive and evasive opportunistic oral pathogen. FEMS Microbiol Lett. 333, 1–9 (2012).
Giacona, M. B. et al. Porphyromonas gingivalis induces its uptake by human macrophages and promotes foam cell formation in vitro. FEMS Microbiol Lett. 241, 95–101 (2004).
Colomer-Winter, C., Gaca, A. O. & Lemos, J. A. Association of Metal Homeostasis and (p)ppGpp Regulation in the Pathophysiology of Enterococcus faecalis. Infect. Immun. 85, https://doi.org/10.1128/IAI.00260-17 (2017).
Mach, H., Hecker, M., Hill, I., Schroeter, A. & Mach, F. [Physiologic significance of “stringent control” in Escherichia coli under extreme starvation]. Z. Naturforsch. C. 44, 838–844 (1989).
Prusa, J., Zhu, D. X. & Stallings, C. L. The stringent response and Mycobacterium tuberculosis pathogenesis. Pathog. Dis. 76, https://doi.org/10.1093/femspd/fty054 (2018).
Roghanian, M., Semsey, S., Lobner-Olesen, A. & Jalalvand, F. (p)ppGpp-mediated stress response induced by defects in outer membrane biogenesis and ATP production promotes survival in Escherichia coli. Sci. Rep. 9, 2934 (2019).
Veith, P. D., Glew, M. D., Gorasia, D. G. & Reynolds, E. C. Type IX secretion: the generation of bacterial cell surface coatings involved in virulence, gliding motility and the degradation of complex biopolymers. Mol. Microbiol 106, 35–53 (2017).
Lasica, A. M., Ksiazek, M., Madej, M. & Potempa, J. The Type IX Secretion System (T9SS): Highlights and Recent Insights into Its Structure and Function. Front. Cell. Infect. Microbiol. 7, 215 (2017).
Moradali, M. F., Ghods, S., Angelini, T. E. & Davey, M. E. Amino acids as wetting agents: surface translocation by Porphyromonas gingivalis. ISME J. https://doi.org/10.1038/s41396-019-0360-9 (2019).
Kadowaki, T. et al. A two-component system regulates gene expression of the type IX secretion component proteins via an ECF sigma factor. Sci. Rep. 6, 23288 (2016).
Ishii, K. et al. Porphyromonas gingivalis peptidoglycans induce excessive activation of the innate immune system in silkworm larvae. J. Biol. Chem. 285, 33338–33347 (2010).
Moye, Z. D., Valiuskyte, K., Dewhirst, F. E., Nichols, F. C. & Davey, M. E. Synthesis of Sphingolipids Impacts Survival of Porphyromonas gingivalis and the Presentation of Surface Polysaccharides. Front. Microbiol. 7, 1919 (2016).
Fletcher, H. M. et al. Virulence of a Porphyromonas gingivalis W83 mutant defective in the prtH gene. Infect. Immun. 63, 1521–1528 (1995).
Gardner, R. G., Russell, J. B., Wilson, D. B., Wang, G. R. & Shoemaker, N. B. Use of a modified Bacteroides-Prevotella shuttle vector to transfer a reconstructed beta-1,4-D-endoglucanase gene into Bacteroides uniformis and Prevotella ruminicola B(1)4. Appl. Environ. Microbiol. 62, 196–202 (1996).
Valentine, P. J., Shoemaker, N. B. & Salyers, A. A. Mobilization of Bacteroides plasmids by Bacteroides conjugal elements. J. Bacteriol. 170, 1319–1324 (1988).
Alberti-Segui, C., Arndt, A., Cugini, C., Priyadarshini, R. & Davey, M. E. HU protein affects transcription of surface polysaccharide synthesis genes in Porphyromonas gingivalis. J. Bacteriol. 192, 6217–6229 (2010).
Scott, J. C., Klein, B. A., Duran-Pinedo, A., Hu, L. & Duncan, M. J. A two-component system regulates hemin acquisition in Porphyromonas gingivalis. PLoS ONE 8, e73351 (2013).
Christopher, A. B., Arndt, A., Cugini, C. & Davey, M. E. A streptococcal effector protein that inhibits Porphyromonas gingivalis biofilm development. Microbiology 156, 3469–3477 (2010).
Kim, H. M., Yoon, C. K., Ham, H. I., Seok, Y. J. & Park, Y. H. Stimulation of Vibrio vulnificus Pyruvate Kinase in the Presence of Glucose to Cope With H2O2 Stress Generated by Its Competitors. Front Microbiol 9, 1112 (2018).
Moye, Z. D., Gormley, C. M. & Davey, M. E. Galactose impacts the size and intracellular composition of the asaccharolytic oral pathobiont Porphyromonas gingivalis. Appl. Environ. Microbiol. 85, https://doi.org/10.1128/AEM.02268-18 (2019).
McClure, R. et al. Computational analysis of bacterial RNA-Seq data. Nucleic Acids Res. 41, e140 (2013).
Park, Y. H., Lee, C. R., Choe, M. & Seok, Y. J. HPr antagonizes the anti-sigma70 activity of Rsd in Escherichia coli. Proc. Natl Acad. Sci. USA 110, 21142–21147 (2013).
Acknowledgements
We would like to thank Dr. Jose A. Lemos (Department of Oral Biology, University of Florida) and Dr. Cristina Colomer Winter (Boehringer Ingelheim, Catalonia, Spain) for providing all the resources needed and technical assistance with assays of (p)ppGpp. We also thank the personnel at the University of Florida, Interdisciplinary Center for Biotechnology Research, and Electron Microscopy Core for their expertise, especially, Karen Kelly. We thank all members of the Davey laboratory for helpful discussions. This work was supported by the National Institute of Dental and Craniofacial Research of the National Institutes of Health under award numbers R01DE019117 and R01DE024580 awarded to M.E.D.
Author information
Authors and Affiliations
Contributions
H.K. contributed to the conception, experimental design, data acquisition, interpretation of data, and paper preparation. M.E.D. contributed to the conception, experimental design, interpretation of data, and editing of the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kim, HM., Davey, M.E. Synthesis of ppGpp impacts type IX secretion and biofilm matrix formation in Porphyromonas gingivalis. npj Biofilms Microbiomes 6, 5 (2020). https://doi.org/10.1038/s41522-020-0115-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41522-020-0115-4
This article is cited by
-
Atypical cyclic di-AMP signaling is essential for Porphyromonas gingivalis growth and regulation of cell envelope homeostasis and virulence
npj Biofilms and Microbiomes (2022)
-
Gain and loss of antibiotic resistant genes in multidrug resistant bacteria: One Health perspective
Journal of Microbiology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.