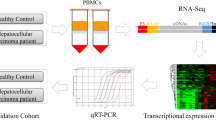
Abstract
Multiple transcript isoforms of genes can be formed by processing and modifying the 5′ and 3′ ends of RNA. Herein, the aim of this study is to uncover the characteristics of RNA processing modification (RPM) in hepatocellular carcinoma (HCC), and to identify novel biomarkers and potential targets for treatment. Firstly, integrated bioinformatics analysis was carried out to identify risk prognostic RPM regulators (RPMRs). Then, we used these RPMRs to identify subtypes of HCC and explore differences in immune microenvironment and cellular function improvement pathways between the sub-types. Finally, we used the principal component analysis algorithms to estimate RPMscore, which were applied to 5 cohorts. Lower RPMscore among patients correlated with a declined survival rate, increased immune infiltration, and raised expression of immune checkpoints, aligning with the “immunity tidal model theory”. The RPMscore exhibited robust, which was validated in multiple datasets. Mechanistically, low RPMscore can create an immunosuppressive microenvironment in HCC by manipulating tumor-associated macrophages. Preclinically, patients with high RPMscore might benefit from immunotherapy. The RPMscore is helpful in clustering HCC patients with distinct prognosis and immunotherapy. Our RPMscore model can help clinicians to select personalized therapy for HCC patients, and RPMscore may act a part in the development of HCC.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 6 digital issues and online access to articles
$119.00 per year
only $19.83 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout











Similar content being viewed by others
Data availability
The datasets generated for this study can be found in the GEO database (GSE14520, GSE36376, GSE76427, GSE20140, GSE27150, GSE140901; https://www.ncbi.nlm.nih.gov/geo/), and UCSC Xena website (https://gdc.xenahubs.net).
References
Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA: a cancer J Clin. 2023;73:17–48.
Vogel A, Meyer T, Sapisochin G, Salem R, Saborowski A. Hepatocellular carcinoma. Lancet (Lond, Engl). 2022;400:1345–62.
Yang JD, Hainaut P, Gores GJ, Amadou A, Plymoth A, Roberts LR. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol. 2019;16:589–604.
Sangro B, Sarobe P, Hervás-Stubbs S, Melero I. Advances in immunotherapy for hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2021;18:525–43.
Llovet JM, Castet F, Heikenwalder M, Maini MK, Mazzaferro V, Pinato DJ, et al. Immunotherapies for hepatocellular carcinoma. Nat Rev Clin Oncol. 2022;19:151–72.
Zhu AX, Finn RS, Edeline J, Cattan S, Ogasawara S, Palmer D, et al. Pembrolizumab in patients with advanced hepatocellular carcinoma previously treated with sorafenib (KEYNOTE-224): a non-randomised, open-label phase 2 trial. Lancet Oncol. 2018;19:940–52.
Gok Yavuz B, Hasanov E, Lee SS, Mohamed YI, Curran MA, Koay EJ, et al. Current Landscape and Future Directions of Biomarkers for Immunotherapy in Hepatocellular Carcinoma. J Hepatocell carcinoma. 2021;8:1195–207.
Ayuso C, Rimola J, Vilana R, Burrel M, Darnell A, García-Criado Á, et al. Diagnosis and staging of hepatocellular carcinoma (HCC): current guidelines. Eur J Radiol. 2018;101:72–81.
Reyes A, Huber W. Alternative start and termination sites of transcription drive most transcript isoform differences across human tissues. Nucleic acids Res. 2018;46:582–92.
Gruber AJ, Zavolan M. Alternative cleavage and polyadenylation in health and disease. Nat Rev Genet. 2019;20:599–614.
Tan S, Zhang M, Shi X, Ding K, Zhao Q, Guo Q, et al. CPSF6 links alternative polyadenylation to metabolism adaption in hepatocellular carcinoma progression. J Exp Clin Cancer Res: CR. 2021;40:85.
Chen SL, Zhu ZX, Yang X, Liu LL, He YF, Yang MM, et al. Cleavage and Polyadenylation Specific Factor 1 Promotes Tumor Progression via Alternative Polyadenylation and Splicing in Hepatocellular Carcinoma. Front cell Dev Biol. 2021;9:616835.
Zeng X, Liao G, Li S, Liu H, Zhao X, Li S, et al. Eliminating METTL1-mediated accumulation of PMN-MDSCs prevents hepatocellular carcinoma recurrence after radiofrequency ablation. Hepatol (Baltim, Md). 2023;77:1122–38.
Zhao S, Ye Z, Stanton R. Misuse of RPKM or TPM normalization when comparing across samples and sequencing protocols. RNA (N.Y, N.Y). 2020;26:903–9.
Parker HS, Leek JT, Favorov AV, Considine M, Xia X, Chavan S, et al. Preserving biological heterogeneity with a permuted surrogate variable analysis for genomics batch correction. Bioinforma (Oxf, Engl). 2014;30:2757–63.
Mariathasan S, Turley SJ, Nickles D, Castiglioni A, Yuen K, Wang Y, et al. TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature. 2018;554:544–8.
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic acids Res. 2015;43:e47.
Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinforma (Oxf, Engl). 2010;26:1572–3.
Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z, et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innov (Camb (Mass)). 2021;2:100141.
Yoshihara K, Shahmoradgoli M, Martínez E, Vegesna R, Kim H, Torres-Garcia W, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612.
Finotello F, Trajanoski Z. Quantifying tumor-infiltrating immune cells from transcriptomics data. Cancer Immunol, Immunother: CII. 2018;67:1031–40.
Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat methods. 2015;12:453–7.
Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18:220.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50.
Su Y, Huang J, Hu J. m(6)A RNA Methylation Regulators Contribute to Malignant Progression and Have Clinical Prognostic Impact in Gastric Cancer. Front Oncol. 2019;9:1038.
Wang X, Zhang S, Han K, Wang L, Liu X. Induction of Apoptosis by Matrine Derivative ZS17 in Human Hepatocellular Carcinoma BEL-7402 and HepG2 Cells through ROS-JNK-P53 Signalling Pathway Activation. Int J Mol Sci. 2022;23:15991.
Li XY, Cui X, Xie CQ, Wu Y, Song T, He JD, et al. Andrographolide causes p53-independent HCC cell death through p62 accumulation and impaired DNA damage repair. Phytomed: Int J Phytother phytopharmacol. 2023;121:155089.
Tang B, Zhu J, Wang Y, Chen W, Fang S, Mao W, et al. Targeted xCT-mediated Ferroptosis and Protumoral Polarization of Macrophages Is Effective against HCC and Enhances the Efficacy of the Anti-PD-1/L1 Response. Adv Sci (Weinh, Baden-Wurtt, Ger). 2023;10:e2203973.
Chen Q, Zheng W, Guan J, Liu H, Dan Y, Zhu L, et al. SOCS2-enhanced ubiquitination of SLC7A11 promotes ferroptosis and radiosensitization in hepatocellular carcinoma. Cell Death Differ. 2023;30:137–51.
Jiang N, Yu Y, Wu D, Wang S, Fang Y, Miao H, et al. HLA and tumour immunology: immune escape, immunotherapy and immune-related adverse events. J Cancer Res Clin Oncol. 2023;149:737–47.
Zhan X, Wu R, Kong XH, You Y, He K, Sun XY, et al. Elevated neutrophil extracellular traps by HBV-mediated S100A9-TLR4/RAGE-ROS cascade facilitate the growth and metastasis of hepatocellular carcinoma. Cancer Commun (Lond, Engl). 2023;43:225–45.
Zhou C, Weng J, Liu C, Liu S, Hu Z, Xie X, et al. Disruption of SLFN11 Deficiency-Induced CCL2 Signaling and Macrophage M2 Polarization Potentiates Anti-PD-1 Therapy Efficacy in Hepatocellular Carcinoma. Gastroenterology. 2023;164:1261–78.
Jiang Y, Hong K, Zhao Y, Xu K. Emerging role of deubiquitination modifications of programmed death-ligand 1 in cancer immunotherapy. Front Immunol. 2023;14:1228200.
Yuan X, Zhou J, Zhou L, Huang Z, Wang W, Qiu J, et al. Apoptosis-Related Gene-Mediated Cell Death Pattern Induces Immunosuppression and Immunotherapy Resistance in Gastric Cancer. Front Genet. 2022;13:921163.
Ma P, Zou C, Xia S. Oncogenic signaling pathway mediated by Notch pathway-related genes induces immunosuppression and immunotherapy resistance in hepatocellular carcinoma. Immunogenetics. 2022;74:539–57.
Zhou L, Xu G, Huang F, Chen W, Zhang J, Tang Y. Apoptosis related genes mediated molecular subtypes depict the hallmarks of the tumor microenvironment and guide immunotherapy in bladder cancer. BMC Med Genomics. 2023;16:88.
Wang S, Chen L, Liu W. Matrix stiffness-dependent STEAP3 coordinated with PD-L2 identify tumor responding to sorafenib treatment in hepatocellular carcinoma. Cancer cell Int. 2022;22:318.
Nie X, Chen W, Zhu Y, Huang B, Yu W, Wu Z, et al. B7-DC (PD-L2) costimulation of CD4(+) T-helper 1 response via RGMb. Cell Mol Immunol. 2018;15:888–97.
Funding
This research was supported by the Construction of Major Subject [Grant no. (YNZDXK202201, 2022-2025)] of Huadu District People´s Hospital of Guangzhou; the construction project of inheritance studio of national famous and old traditional Chinese Medicine experts (Grant no.140000020132).
Author information
Authors and Affiliations
Contributions
M.D. and X.H.L. conceived the project. S.L. acquired and processed raw sequencing data. L.B.Z. performed data integration and conducted data analysis. X.H.L. and S.L. assisted in the interpretation of results. C.B.Z. wrote the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
The study was approved by the ethical committee in the Renmin Hospital, Hubei University of Medicine, and conducted under the guidance of the Declaration of Helsinki.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, X., Liu, S., Zou, L. et al. RNA processing modification mediated subtypes illustrate the distinctive features of tumor microenvironment in hepatocellular carcinoma. Genes Immun 25, 132–148 (2024). https://doi.org/10.1038/s41435-024-00265-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41435-024-00265-8