Abstract
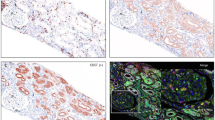
Interferon-γ (IFN-γ) is an important cytokine in tissue homeostasis and immune response, while studies about it in antibody-mediated rejection (ABMR) are very limited. This study aims to comprehensively elucidate the role of IFN-γ in ABMR after renal transplantation. In six renal transplantation cohorts, the IFN-γ responses (IFNGR) biological process was consistently top up-regulated in ABMR compared to stable renal function or even T cell-mediated rejection in both allografts and peripheral blood. According to single-cell analysis, IFNGR levels were found to be broadly elevated in most cell types in allografts and peripheral blood with ABMR. In allografts with ABMR, M1 macrophages had the highest IFNGR levels and were heavily infiltrated, while kidney resident M2 macrophages were nearly absent. In peripheral blood, CD14+ monocytes had the top IFNGR level and were significantly increased in ABMR. Immunofluorescence assay showed that levels of IFN-γ and M1 macrophages were sharply elevated in allografts with ABMR than non-rejection. Importantly, the IFNGR level in allografts was identified as a strong risk factor for long-term renal graft survival. Together, this study systematically analyzed multi-omics from thirteen independent cohorts and identified IFN-γ and IFNGR as determinants of ABMR and clinical outcomes in patients after renal transplantation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 6 digital issues and online access to articles
$119.00 per year
only $19.83 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout











Similar content being viewed by others
Data availability
The data used to support the findings of this study are available from the corresponding author upon request.
References
Wolfe RA, Ashby VB, Milford EL, Ojo AO, Ettenger RE, Agodoa LY, et al. Comparison of mortality in all patients on dialysis, patients on dialysis awaiting transplantation, and recipients of a first cadaveric transplant. N Engl J Med. 1999;341:1725–30.
Hariharan S, Israni AK, Danovitch G. Long-Term Survival after Kidney Transplantation. N Engl J Med. 2021;385:729–43.
Einecke G, Sis B, Reeve J, Mengel M, Campbell PM, Hidalgo LG, et al. Antibody-mediated microcirculation injury is the major cause of late kidney transplant failure. Am J Transpl. 2009;9:2520–31.
Willicombe M, Roufosse C, Brookes P, Galliford JW, McLean AG, Dorling A, et al. Antibody-mediated rejection after alemtuzumab induction: incidence, risk factors, and predictors of poor outcome. Transplantation. 2011;92:176–82.
Lucas JG, Co JP, Nwaogwugwu UT, Dosani I, Sureshkumar KK. Antibody-mediated rejection in kidney transplantation: an update. Expert Opin Pharm. 2011;12:579–92.
Schinstock CA, Mannon RB, Budde K, Chong AS, Haas M, Knechtle S, et al. Recommended Treatment for Antibody-mediated Rejection After Kidney Transplantation: The 2019 Expert Consensus From the Transplantion Society Working Group. Transplantation. 2020;104:911–22.
Velidedeoglu E, Cavaillé-Coll MW, Bala S, Belen OA, Wang Y, Albrecht R. Summary of 2017 FDA Public Workshop: Antibody-mediated Rejection in Kidney Transplantation. Transplantation. 2018;102:e257–64.
Burton SA, Amir N, Asbury A, Lange A, Hardinger KL. Treatment of antibody-mediated rejection in renal transplant patients: a clinical practice survey. Clin Transpl. 2015;29:118–23.
Sautenet B, Blancho G, Büchler M, Morelon E, Toupance O, Barrou B, et al. One-year Results of the Effects of Rituximab on Acute Antibody-Mediated Rejection in Renal Transplantation: RITUX ERAH, a Multicenter Double-blind Randomized Placebo-controlled Trial. Transplantation. 2016;100:391–9.
Eskandary F, Regele H, Baumann L, Bond G, Kozakowski N, Wahrmann M, et al. A Randomized Trial of Bortezomib in Late Antibody-Mediated Kidney Transplant Rejection. J Am Soc Nephrol. 2018;29:591–605.
Cornell LD, Schinstock CA, Gandhi MJ, Kremers WK, Stegall MD. Positive crossmatch kidney transplant recipients treated with eculizumab: outcomes beyond 1 year. Am J Transpl. 2015;15:1293–302.
Gill RG, Lin CM. Linking innate immunity and chronic antibody-mediated allograft rejection. Curr Opin Organ Tran. 2019;24:694–8.
Usuelli V, Loretelli C, Seelam AJ, Pastore I, D’Addio F, Ben NM, et al. Novel Soluble Mediators of Innate Immune System Activation in Solid Allograft Rejection. Transplantation. 2022;106:500–9.
Ivashkiv LB. IFNγ: signalling, epigenetics and roles in immunity, metabolism, disease and cancer immunotherapy. Nat Rev Immunol. 2018;18:545–58.
Hu X, Ivashkiv LB. Cross-regulation of signaling pathways by interferon-gamma: implications for immune responses and autoimmune diseases. Immunity. 2009;31:539–50.
Boehm U, Klamp T, Groot M, Howard JC. Cellular responses to interferon-gamma. Annu Rev Immunol. 1997;15:749–95.
McLaren JE, Ramji DP. Interferon gamma: a master regulator of atherosclerosis. Cytokine Growth F R. 2009;20:125–35.
Famulski KS, Einecke G, Reeve J, Ramassar V, Allanach K, Mueller T, et al. Changes in the transcriptome in allograft rejection: IFN-gamma-induced transcripts in mouse kidney allografts. Am J Transpl. 2006;6:1342–54.
Kong D, Huang S, Miao X, Li J, Wu Z, Shi Y, et al. The dynamic cellular landscape of grafts with acute rejection after heart transplantation. J Heart Lung Transpl. 2023;42:160–72.
Reeve J, Böhmig GA, Eskandary F, Einecke G, Lefaucheur C, Loupy A, et al. Assessing rejection-related disease in kidney transplant biopsies based on archetypal analysis of molecular phenotypes. JCI Insight. 2017;2:e94197.
Reeve J, Sellarés J, Mengel M, Sis B, Skene A, Hidalgo L, et al. Molecular diagnosis of T cell-mediated rejection in human kidney transplant biopsies. Am J Transpl. 2013;13:645–55.
Halloran PF, Pereira AB, Chang J, Matas A, Picton M, De Freitas D, et al. Potential impact of microarray diagnosis of T cell-mediated rejection in kidney transplants: The INTERCOM study. Am J Transpl. 2013;13:2352–63.
Broin P Ó, Hayde N, Bao Y, Ye B, Calder RB, de Boccardo G, et al. A pathogenesis-based transcript signature in donor-specific antibody-positive kidney transplant patients with normal biopsies. Genom Data. 2014;2:357–60.
Van Loon E, Lamarthée B, de Loor H, Van Craenenbroeck AH, Brouard S, Danger R, et al. Biological pathways and comparison with biopsy signals and cellular origin of peripheral blood transcriptomic profiles during kidney allograft pathology. Kidney Int. 2022;102:183–95.
Van Loon E, Gazut S, Yazdani S, Lerut E, de Loor H, Coemans M, et al. Development and validation of a peripheral blood mRNA assay for the assessment of antibody-mediated kidney allograft rejection: A multicentre, prospective study. Ebiomedicine. 2019;46:463–72.
Einecke G, Reeve J, Sis B, Mengel M, Hidalgo L, Famulski KS, et al. A molecular classifier for predicting future graft loss in late kidney transplant biopsies. J Clin Invest. 2010;120:1862–72.
Zhang W, Yi Z, Wei C, Keung KL, Sun Z, Xi C, et al. Pretransplant transcriptomic signature in peripheral blood predicts early acute rejection. JCI Insight. 2019;4:e127543.
Hrubá P, Brabcová I, Gueler F, Krejčík Z, Stránecký V, Svobodová E, et al. Molecular diagnostics identifies risks for graft dysfunction despite borderline histologic changes. Kidney Int. 2015;88:785–95.
Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27:1739–40.
Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453–7.
Hu FF, Liu CJ, Liu LL, Zhang Q, Guo AY. Expression profile of immune checkpoint genes and their roles in predicting immunotherapy response. Brief Bioinform. 2021;22:bbaa176.
Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12:252–64.
Campbell KS, Purdy AK. Structure/function of human killer cell immunoglobulin-like receptors: lessons from polymorphisms, evolution, crystal structures and mutations. Immunology. 2011;132:315–25.
Afrache H, Gouret P, Ainouche S, Pontarotti P, Olive D. The butyrophilin (BTN) gene family: from milk fat to the regulation of the immune response. Immunogenetics. 2012;64:781–94.
Suryawanshi H, Yang H, Lubetzky M, Morozov P, Lagman M, Thareja G, et al. Detection of infiltrating fibroblasts by single-cell transcriptomics in human kidney allografts. Plos One. 2022;17:e0267704.
Kong F, Ye S, Zhong Z, Zhou X, Zhou W, Liu Z, et al. Single-Cell Transcriptome Analysis of Chronic Antibody-Mediated Rejection After Renal Transplantation. Front Immunol. 2021;12:767618.
Hao Y, Hao S, Andersen-Nissen E, Mauck WR, Zheng S, Butler A, et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184:3573–87.
Korsunsky I, Millard N, Fan J, Slowikowski K, Zhang F, Wei K, et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods. 2019;16:1289–96.
Zheng L, Qin S, Si W, Wang A, Xing B, Gao R, et al. Pan-cancer single-cell landscape of tumor-infiltrating T cells. Science. 2021;374:abe6474.
Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014;32:381–6.
Jin S, Guerrero-Juarez CF, Zhang L, Chang I, Ramos R, Kuan CH, et al. Inference and analysis of cell-cell communication using CellChat. Nat Commun. 2021;12:1088.
De Benedetti F, Prencipe G, Bracaglia C, Marasco E, Grom AA. Targeting interferon-γ in hyperinflammation: opportunities and challenges. Nat Rev Rheumatol. 2021;17:678–91.
Hidalgo LG, Sis B, Sellares J, Campbell PM, Mengel M, Einecke G, et al. NK cell transcripts and NK cells in kidney biopsies from patients with donor-specific antibodies: evidence for NK cell involvement in antibody-mediated rejection. Am J Transpl. 2010;10:1812–22.
Montgomery RA, Zachary AA, Racusen LC, Leffell MS, King KE, Burdick J, et al. Plasmapheresis and intravenous immune globulin provides effective rescue therapy for refractory humoral rejection and allows kidneys to be successfully transplanted into cross-match-positive recipients. Transplantation. 2000;70:887–95.
Rocha PN, Butterly DW, Greenberg A, Reddan DN, Tuttle-Newhall J, Collins BH, et al. Beneficial effect of plasmapheresis and intravenous immunoglobulin on renal allograft survival of patients with acute humoral rejection. Transplantation. 2003;75:1490–5.
Böhmig GA, Wahrmann M, Regele H, Exner M, Robl B, Derfler K, et al. Immunoadsorption in severe C4d-positive acute kidney allograft rejection: a randomized controlled trial. Am J Transpl. 2007;7:117–21.
Lefaucheur C, Nochy D, Andrade J, Verine J, Gautreau C, Charron D, et al. Comparison of combination Plasmapheresis/IVIg/anti-CD20 versus high-dose IVIg in the treatment of antibody-mediated rejection. Am J Transpl. 2009;9:1099–107.
Loupy A, Lefaucheur C. Antibody-Mediated Rejection of Solid-Organ Allografts. N Engl J Med. 2018;379:1150–60.
Asderakis A, Sankaran D, Dyer P, Johnson RW, Pravica V, Sinnott PJ, et al. Association of polymorphisms in the human interferon-gamma and interleukin-10 gene with acute and chronic kidney transplant outcome: the cytokine effect on transplantation. Transplantation. 2001;71:674–7.
Awad M, Pravica V, Perrey C, El GA, Yonan N, Sinnott PJ, et al. CA repeat allele polymorphism in the first intron of the human interferon-gamma gene is associated with lung allograft fibrosis. Hum Immunol. 1999;60:343–6.
Jackson SW, Jacobs HM, Arkatkar T, Dam EM, Scharping NE, Kolhatkar NS, et al. B cell IFN-γ receptor signaling promotes autoimmune germinal centers via cell-intrinsic induction of BCL-6. J Exp Med. 2016;213:733–50.
Domeier PP, Chodisetti SB, Soni C, Schell SL, Elias MJ, Wong EB, et al. IFN-γ receptor and STAT1 signaling in B cells are central to spontaneous germinal center formation and autoimmunity. J Exp Med. 2016;213:715–32.
Loupy A, Haas M, Roufosse C, Naesens M, Adam B, Afrouzian M, et al. The Banff 2019 Kidney Meeting Report (I): Updates on and clarification of criteria for T cell- and antibody-mediated rejection. Am J Transpl. 2020;20:2318–31.
Sablik KA, Jordanova ES, Pocorni N, Clahsen-van GM, Betjes M. Immune Cell Infiltrate in Chronic-Active Antibody-Mediated Rejection. Front Immunol. 2019;10:3106.
Lin SL, Li B, Rao S, Yeo EJ, Hudson TE, Nowlin BT, et al. Macrophage Wnt7b is critical for kidney repair and regeneration. P Natl Acad Sci USA. 2010;107:4194–9.
Schmidt IM, Hall IE, Kale S, Lee S, He CH, Lee Y, et al. Chitinase-like protein Brp-39/YKL-40 modulates the renal response to ischemic injury and predicts delayed allograft function. J Am Soc Nephrol. 2013;24:309–19.
Sola A, Weigert A, Jung M, Vinuesa E, Brecht K, Weis N, et al. Sphingosine-1-phosphate signalling induces the production of Lcn-2 by macrophages to promote kidney regeneration. J Pathol. 2011;225:597–608.
Callemeyn J, Lamarthée B, Koenig A, Koshy P, Thaunat O, Naesens M. Allorecognition and the spectrum of kidney transplant rejection. Kidney Int. 2022;101:692–710.
Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol. 2008;8:958–69.
Panzer SE. Macrophages in Transplantation: A Matter of Plasticity, Polarization, and Diversity. Transplantation. 2022;106:257–67.
Schinstock CA, Stegall M, Cosio F. New insights regarding chronic antibody-mediated rejection and its progression to transplant glomerulopathy. Curr Opin Nephrol Hy. 2014;23:611–8.
Toki D, Zhang W, Hor KL, Liuwantara D, Alexander SI, Yi Z, et al. The role of macrophages in the development of human renal allograft fibrosis in the first year after transplantation. Am J Transpl. 2014;14:2126–36.
Nash WT, Okusa MD. Chess Not Checkers: Complexities Within the Myeloid Response to the Acute Kidney Injury Syndrome. Front Med-Lausanne. 2021;8:676688.
Uderhardt S, Martins AJ, Tsang JS, Lämmermann T, Germain RN. Resident Macrophages Cloak Tissue Microlesions to Prevent Neutrophil-Driven Inflammatory Damage. Cell. 2019;177:541–55.
Narasimhan PB, Marcovecchio P, Hamers A, Hedrick CC. Nonclassical Monocytes in Health and Disease. Annu Rev Immunol. 2019;37:439–56.
Ng CT, Fong LY, Sulaiman MR, Moklas MA, Yong YK, Hakim MN, et al. Interferon-Gamma Increases Endothelial Permeability by Causing Activation of p38 MAP Kinase and Actin Cytoskeleton Alteration. J Inter Cytok Res. 2015;35:513–22.
Stolpen AH, Guinan EC, Fiers W, Pober JS. Recombinant tumor necrosis factor and immune interferon act singly and in combination to reorganize human vascular endothelial cell monolayers. Am J Pathol. 1986;123:16–24.
Osickova K, Hruba P, Kabrtova K, Klema J, Maluskova J, Slavcev A, et al. Predictive Potential of Flow Cytometry Crossmatching in Deceased Donor Kidney Transplant Recipients Subjected to Peritransplant Desensitization. Front Med Lausanne. 2021;8:780636.
Turgeon NA, Kirk AD, Iwakoshi NN. Differential effects of donor-specific alloantibody. Transpl Rev Orlan. 2009;23:25–33.
Viglietti D, Lefaucheur C, Glotz D. Evidence for an important role of both complement-binding and noncomplement-binding donor-specific antibodies in renal transplantation. Curr Opin Organ Tran. 2016;21:433–40.
El-Awar N, Jucaud V, Nguyen A. HLA Epitopes: The Targets of Monoclonal and Alloantibodies Defined. J Immunol Res. 2017;2017:3406230.
Duquesnoy RJ. HLA epitope based matching for transplantation. Transpl Immunol. 2014;31:1–6.
Konvalinka A, Tinckam K. Utility of HLA Antibody Testing in Kidney Transplantation. J Am Soc Nephrol. 2015;26:1489–502.
Tait BD. Detection of HLA Antibodies in Organ Transplant Recipients - Triumphs and Challenges of the Solid Phase Bead Assay. Front Immunol. 2016;7:570.
Buatois V, Chatel L, Cons L, Lory S, Richard F, Guilhot F, et al. Use of a mouse model to identify a blood biomarker for IFNγ activity in pediatric secondary hemophagocytic lymphohistiocytosis. Transl Res. 2017;180:37–52.
Lortat-Jacob H, Baltzer F, Grimaud JA. Heparin decreases the blood clearance of interferon-gamma and increases its activity by limiting the processing of its carboxyl-terminal sequence. J Biol Chem. 1996;271:16139–43.
Rutenfranz I, Kirchner H. Pharmacokinetics of recombinant murine interferon-gamma in mice. J Interferon Res. 1988;8:573–80.
Locatelli F, Jordan MB, Allen C, Cesaro S, Rizzari C, Rao A, et al. Emapalumab in Children with Primary Hemophagocytic Lymphohistiocytosis. N. Engl J Med. 2020;382:1811–22.
Hommes DW, Mikhajlova TL, Stoinov S, Stimac D, Vucelic B, Lonovics J, et al. Fontolizumab, a humanised anti-interferon gamma antibody, demonstrates safety and clinical activity in patients with moderate to severe Crohn’s disease. Gut. 2006;55:1131–7.
Reinisch W, de Villiers W, Bene L, Simon L, Rácz I, Katz S, et al. Fontolizumab in moderate to severe Crohn’s disease: a phase 2, randomized, double-blind, placebo-controlled, multiple-dose study. Inflamm Bowel Dis. 2010;16:233–42.
Boedigheimer MJ, Martin DA, Amoura Z, Sánchez-Guerrero J, Romero-Diaz J, Kivitz A, et al. Safety, pharmacokinetics and pharmacodynamics of AMG 811, an anti-interferon-γ monoclonal antibody, in SLE subjects without or with lupus nephritis. Lupus Sci Med. 2017;4:e000226.
Chen P, Vu T, Narayanan A, Sohn W, Wang J, Boedigheimer M, et al. Pharmacokinetic and pharmacodynamic relationship of AMG 811, an anti-IFN-γ IgG1 monoclonal antibody, in patients with systemic lupus erythematosus. Pharm Res Dordr. 2015;32:640–53.
Kobayashi T, Hibi T. Improving IBD outcomes in the era of many treatment options. Nat Rev Gastro Hepat. 2023;20:79–80.
Funding
This work was supported by the General Program of the National Natural Science Foundation of China (NSFC) (81970645, 82000711, 82170766) and the joint fund of the Beijing Municipal Commission of Education and Natural Science Foundation of Beijing Municipality (KZ202010025036).
Author information
Authors and Affiliations
Contributions
All authors contributed to the manuscript. Hao Zhang, DZ, ZZ, and XH designed the study. Hao Zhang and YX performed the bulk data analysis and DZ conducted the single-cell data analysis. Hao Zhang and DZ drew the figures. Hao Zhang and He Zhang collected the patient information. DZ conducted the immunofluorescence staining experiment. Hao Zhang drafted the manuscript. YX, DZ, ZZ, and XH revised the manuscript. YX provided clinical suggestions and feedback for the study. All authors have read and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
All procedures were performed strictly in compliance with the Declaration of Helsinki 1964 or equivalent ethical principles. The research protocol was approved by the Ethics Committee of Beijing Chao-Yang Hospital (Approval number 2023-科-16). All patients involved in this study had given their informed consent before the study.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, H., Zhang, D., Xu, Y. et al. Interferon-γ and its response are determinants of antibody-mediated rejection and clinical outcomes in patients after renal transplantation. Genes Immun 25, 66–81 (2024). https://doi.org/10.1038/s41435-024-00254-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41435-024-00254-x