Abstract
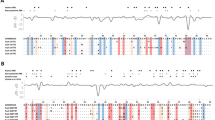
Functional diversification, a higher evolutionary rate, and intense positive selection help a limited number of immune genes interact with many pathogens. Repeats in protein-coding regions are a well-known source of functional diversification, adaptive variation, and evolutionary novelty in a short time. Repeats play a crucial role in biochemical functions like functional diversification of transcription regulation, protein kinases, cell adhesion, signaling pathways, morphogenesis, DNA repair, recombination, and RNA processing. Repeat length variation can change the associated protein’s interaction, efficacy, and overall protein network. Repeats have an intrinsic unstable nature and can potentially evolve rapidly and expedite the acquisition of complex phenotypic traits and functions. Because of their ability to generate rapid, adaptive variations over short evolutionary distances, repeats are considered “tuning knobs.” Repeat length variation in specific genes, like RUNX2 and ALX4, is associated with morphological and physiological changes across vertebrates. Here we study repeat length variation as a potent source of species-specific immune diversification across several clades of tetrapods. Moreover, we provide a clade-wise comprehensive list of immune genes with repeat types for future studies of morphological/evolutionary changes within species groups. We observe significant repeat length variation of FASLG and C1QC in Rodentia and Primates’ contrasting species groups, respectively.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 6 digital issues and online access to articles
$119.00 per year
only $19.83 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All data associated with this study are available in the Supplementary Materials, and data and scripts used for analysis are provided in an easy-to-browse format: https://github.com/ceglablokdeep/Immune_genes_repeats. A copy of the data has been uploaded on Mendeley datasets with https://doi.org/10.17632/8zxtjh8fjs.1.
References
Shultz AJ, Sackton TB. Immune genes are hotspots of shared positive selection across birds and mammals. Elife. 2019;8:e41815.
Enard D, Cai L, Gwennap C, Petrov DA. Viruses are a dominant driver of protein adaptation in mammals. Elife. 2016;5:e12469.
Kosiol C, Vinar T, Da Fonseca RR, Hubisz MJ, Bustamante CD, Nielsen R, et al. Patterns of positive selection in six Mammalian genomes. PLoS Genet. 2008;4:e1000144.
Alcaide M, Edwards SV. Molecular evolution of the toll-like receptor multigene family in birds. Mol Biol Evol. 2011;28:1703–15.
Carpentier KS, Geballe AP. An evolutionary view of the arms race between protein kinase R and large DNA viruses. J Virol. 2016;90:3280–3.
Shaw AE, Hughes J, Gu Q, Behdenna A, Singer JB, Dennis T, et al. Fundamental properties of the mammalian innate immune system revealed by multispecies comparison of type I interferon responses. PLoS Biol. 2017;15:e2004086.
Solbakken MH, Rise ML, Jakobsen KS, Jentoft S. Successive losses of central immune genes characterize the Gadiformes’ alternate immunity. Genome Biol Evol. 2016;8:3508–15.
Blumer M, Brown T, Freitas MB, Destro AL, Oliveira JA, Morales AE, et al. Gene losses in the common vampire batilluminate molecular adaptations to blood feeding. Sci Adv. 2022;8:eabm6494.
Zimmer MM, Kibe A, Rand U, Pekarek L, Ye L, Buck S, et al. The short isoform of the host antiviral protein ZAP acts as an inhibitor of SARS-CoV-2 programmed ribosomal frameshifting. Nat Commun. 2021;12:1–15.
Yang L, Fu J, Zhou Y. Circular RNAs and their emerging roles in immune regulation. Front Immunol. 2018;9:2977.
Vierbuchen T, Fitzgerald KA. Long non-coding RNAs in antiviral immunity. Semin Cell Dev Biol. 2021;111:126–34.
Persi E, Wolf YI, Koonin EV. Positive and strongly relaxed purifying selection drive the evolution of repeats in proteins. Nat Commun.2016;7:13570.
Marcotte EM, Pellegrini M, Yeates TO, Eisenberg D. A census of protein repeats. J Mol Biol. 1999;293:151–60.
Albà MM, Tompa P, Veitia RA. Amino acid repeats and the structure and evolution of proteins. Gene and Protein Evolution. KARGER: Basel. 2007;3:119–30.
Persi E, Horn D. Systematic analysis of compositional order of proteins reveals new characteristics of biological functions and a universal correlate of macroevolution. PLoS Comput Biol. 2013;9:e1003346.
Andrade MA, Perez-Iratxeta C, Ponting CP. Protein repeats: structures, functions, and evolution. J Struct Biol. 2001;134:117–31.
Karlin S, Brocchieri L, Bergman A, Mrázek J, Gentles AJ. Amino acid runs in eukaryotic proteomes and disease associations. Proc Natl Acad Sci. 2002;99:333–8.
Pellegrini M, Marcotte EM, Yeates TO. A fast algorithm for genome-wide analysis of proteins with repeated sequences. Proteins Struct Funct Genet. 1999;35:440–6.
Kashi Y, King DG. Simple sequence repeats as advantageous mutators in evolution. Trends Genet. 2006;22:253–9.
King DG, Soller M, Kashi Y. Evolutionary tuning knobs. Endeavour. 1997;21:36–40.
Lynch VJ, Wagner GP. Resurrecting the role of transcription factor change in developmental evolution. Evolution (N. Y). 2008;62:2131–54.
Hancock JM, Simon M. Simple sequence repeats in proteins and their significance for network evolution. Gene. 2005;345:113–8.
Myers KA, Rahi-Saund V, Davison MD, Young JA, Cheater AJ, Stern PL. Isolation of a cDNA encoding 5T4 oncofetal trophoblast glycoprotein. An antigen associated with metastasis contains leucine-rich repeats. J Biol Chem. 1994;269:9319–24.
Eldon E, Kooyer S, D’Evelyn D, Duman M, Lawinger P, Botas J, et al. The Drosophila 18 wheeler is required for morphogenesis and has striking similarities to Toll. Development. 1994;120:885–99.
Kobe B, Deisenhofer J. Proteins with leucine-rich repeats. Curr Opin Struct Biol. 1995;5:409–16.
Huntley M, Golding GB. Evolution of simple sequence in proteins. J Mol Evol. 2000;51:131–40.
Gemayel R, Vinces MD, Legendre M, Verstrepen KJ. Variable tandem repeats accelerate evolution of coding and regulatory sequences. 2010. https://doi.org/10.1146/annurev-genet-072610-155046.
Fondon JW, Garner HR. Molecular origins of rapid and continuous morphological evolution. Proc Natl Acad Sci USA. 2004;101:18058–63.
Pajic P, Shen S, Qu J, May AJ, Knox S, Ruhl S, et al. A mechanism of gene evolution generating mucin function. Sci Adv. 2022;8. https://doi.org/10.1126/SCIADV.ABM8757.
Björklund ÅK, Ekman D, Elofsson A. Expansion of protein domain repeats. PLoS Comput Biol. 2006;2:0959–70.
Newton AH, Pask AJ. Evolution and expansion of the RUNX2 QA repeat corresponds with the emergence of vertebrate complexity. Commun Biol. 2020;3:771.
Fondon JW, Garner HR. Molecular origins of rapid and continuous morphological evolution. Proc Natl Acad Sci USA. 2004;101:18058–63.
Mevel R, Draper JE, Lie-a-Ling M, Kouskoff V, Lacaud G. RUNX transcription factors: orchestrators of development. Development. 2019;146:1–19.
Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell. 1997;89:747–54.
Gaillard JL, Berche P, Frehel C, Gouln E, Cossart P. Entry of L. monocytogenes into cells is mediated by internalin, a repeat protein reminiscent of surface antigens from gram-positive cocci. Cell. 1991;65:1127–41.
Kȩdzierski Ł, Montgomery J, Curtis J, Handman E. Leucine-rich repeats in host-pathogen interactions. Arch Immunol Ther Exp (Warsz). 2004;52:104–12.
Reeder JC, Brown GV. Antigenic variation and immune evasion in Plasmodium falciparum malaria. Immunol Cell Biol. 1996;74:546–54.
Schofield L. On the function of repetitive domains in protein antigens of Plasmodium and other eukaryotic parasites. Parasitol Today. 1991;7:99–105.
Cohn M. The immune system: a weapon of mass destruction invented by evolution to even the odds during the war of the DNAs. Immunol Rev. 2002;185:24–38.
Sieling PA, Modlin RL. Toll-like receptors: Mammalian ‘taste receptors’ for a smorgasbord of microbial invaders. Curr Opin Microbiol. 2002;5:70–5.
Medzhitov R. Toll-like receptors and innate immunity. Nat Rev Immunol. 2001;1:135–45.
Cunningham F, Allen JE, Allen J, Alvarez-Jarreta J, Amode MR, Armean IM, et al. Ensembl 2022. Nucleic Acids Res. 2022;50:D988–95.
Agarwala R, Barrett T, Beck J, Benson DA, Bollin C, Bolton E, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2016;44:D7.
Kumar S, Stecher G, Suleski M, Hedges SB. TimeTree: a resource for timelines, timetrees, and divergence times. Mol Biol Evol. 2017;34:1812–9.
Jetz W, Thomas GH, Joy JB, Hartmann K, Mooers AO. The global diversity of birds in space and time. Nature. 2012;491:444–8.
Harrison PM. fLPS 2.0: rapid annotation of compositionally-biased regions in biological sequences. PeerJ. 2021;9:e12363.
Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113.
Revell LJ. phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol Evol. 2012;3:217–23.
R Core Team. R: a language and environment for statistical computing. 2021. https://www.r-project.org.
Smith MD, Wertheim JO, Weaver S, Murrell B, Scheffler K, Kosakovsky Pond SL. Less is more: an adaptive branch-site random effects model for efficient detection of episodic diversifying selection. Mol Biol Evol. 2015;32:1342–53.
Kosakovsky Pond SL, Frost SDW, Muse SV. HyPhy: hypothesis testing using phylogenies. Bioinformatics. 2005;21:676–79.
Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–91.
Ge SX, Jung D, Jung D, Yao R. ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinformatics. 2020;36:2628–9.
Montgomerie S, Cruz JA, Shrivastava S, Arndt D, Berjanskii M, Wishart DS. PROTEUS2: a web server for comprehensive protein structure prediction and structure-based annotation. Nucleic Acids Res. 2008;36:W202–9.
Omasits U, Ahrens CH, Müller S, Wollscheid B. Protter: interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics. 2014;30:884–6.
Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–9.
Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–303.
Goddard TD, Huang CC, Meng EC, Pettersen EF, Couch GS, Morris JH, et al. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 2018;27:14–25.
Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–2.
Deryusheva EI, Machulin AV, Galzitskaya OV. Structural, functional, and evolutionary characteristics of proteins with repeats. Mol Biol. 2021;55:683–704.
Janeway CA. Approaching the asymptote? Evolution and revolution in immunology. J Immunol. 2013;191:4475–87.
Medzhitov R. Recognition of microorganisms and activation of the immune response. Nature. 2007;449:819–26.
Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801.
Inohara N, Chamaillard M, McDonald C, Nuñez G. NOD-LRR proteins: role in host-microbial interactions and inflammatory disease. Annu Rev Biochem. 2005;74:355–83.
Meylan E, Tschopp J, Karin M. Intracellular pattern recognition receptors in the host response. Nature. 2006;442:39–44.
Schatz DG, Oettinger MA, Schlissel MS. V (D) J RECOMBINATION: molecular biology and regulation. Annu Rev Immunol. 1992;10:359–83.
Cocquet J, De Baere E, Caburet S, Veitia RA. Compositional biases and polyalanine runs in humans. Genetics. 2003;165:1613–7.
Albà MM, Guigó R. Comparative analysis of amino acid repeats in rodents and humans. Genome Res. 2004;14:549–54.
Mier P, Alanis-Lobato G, Andrade-Navarro MA. Context characterization of amino acid homorepeats using evolution, position, and order. Proteins Struct Funct Bioinforma. 2017;85:709–19.
Jorda J, Xue B, Uversky VN, Kajava AV. Protein tandem repeats – the more perfect, the less structured. FEBS J. 2010;277:2673–82.
Huntley MA, Golding GB. Simple sequences are rare in the Protein Data Bank. Proteins Struct Funct Genet. 2002;48:134–40.
Wright PE, Dyson HJ. Intrinsically unstructured proteins: Re-assessing the protein structure-function paradigm. J Mol Biol. 1999;293:321–31.
Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6:197–208.
Tompa P. The interplay between structure and function in intrinsically unstructured proteins. FEBS Lett. 2005;579:3346–54.
Dosztányi Z, Chen J, Dunker AK, Simon I, Tompa P. Disorder and sequence repeats in hub proteins and their implications for network evolution. J Proteome Res. 2006;5:2985–95.
Tompa P. Intrinsically unstructured proteins evolve by repeat expansion. BioEssays. 2003;25:847–55.
Chavali S, Chavali PL, Chalancon G, de Groot NS, Gemayel R, Latysheva NS, et al. Constraints and consequences of the emergence of amino acid repeats in eukaryotic proteins. Nat Struct Mol Biol. 2017;24:765–77.
Jones AL, Hulett MD, Parish CR. Histidine-rich glycoprotein: a novel adaptor protein in plasma that modulates the immune, vascular and coagulation systems. Immunol Cell Biol. 2005;83:106–18.
Koide T. Amino acid sequence of human histidine-rich glycoprotein derived from the nucleotide sequence of its cDNA. Biochemistry. 1986;25:2212–20.
Morgan WT. Interactions of the Histidine-Rich glycoprotein of serum with metals. Biochemistry. 1981;20:1054–61.
Hawash MBF, Sanz-Remón J, Grenier JC, Kohn J, Yotova V, Johnson Z, et al. Primate innate immune responses to bacterial and viral pathogens reveals an evolutionary trade-off between strength and specificity. Proc Natl Acad Sci USA. 2021;118:1–10.
Hancock JM, Worthey EA, Santibáñez-Koref MF. A role for selection in regulating the evolutionary emergence of disease-causing and other coding CAG repeats in humans and mice. Mol Biol Evol. 2001;18:1014–23.
Pelletier N, Champagne N, Stifani S, Yang XJ. MOZ and MORF histone acetyltransferases interact with the Runt-domain transcription factor Runx2. Oncogene. 2002;21:2729–40.
Komori T. Regulation of bone development and maintenance by Runx2. Front Biosci. 2008;13:898.
Nagata S, Golstein P. The Fas death factor. Science (80-). 1995;267:1449–56.
Ashkenazi A, Dixit VM. Death receptors: signaling and modulation. Science (80-). 1998;281:1305–8.
Li Y, Sun Y, Cai M, Zhang H, Gao N, Huang H, et al. Fas ligand gene (Faslg) plays an important role in nerve degeneration and regeneration after rat sciatic nerve injury. Front Mol Neurosci. 2018;11:1–13.
Sun M, Lee S, Karray S, Levi-Strauss M, Ames KT, Fink PJ. Cutting Edge: two distinct Motifs within the Fas ligand tail regulate Fas ligand-mediated costimulation. J Immunol. 2007;179:5639–43.
Blott EJ, Bossi G, Clark R, Zvelebil M, Griffiths GM. Fas ligand is targeted to secretory lysosomes via a proline-rich domain in its cytoplasmic tail. J Cell Sci. 2001;114:2405–16.
McCord JP, Grove TZ. Engineering repeat proteins of the immune system. Biopolymers. 2020;111. https://doi.org/10.1002/bip.23348.
Zhang M, Wang F, Li S, Wang Y, Bai Y, Xu X. TALE: a tale of genome editing. Prog Biophys Mol Biol. 2014;114:25–32.
Kelley J, De Bono B, Trowsdale J. IRIS: a database surveying known human immune system genes. Genomics. 2005;85:503–11.
Abeler-Dörner L, Laing AG, Lorenc A, Ushakov DS, Clare S, Speak AO, et al. High-throughput phenotyping reveals expansive genetic and structural underpinnings of immune variation. Nat Immunol. 2020;21:86–100.
Rusinova I, Forster S, Yu S, Kannan A, Masse M, Cumming H, et al. INTERFEROME v2.0: an updated database of annotated interferon-regulated genes. Nucleic Acids Res. 2013;41:D1040-6.
Acknowledgements
We thank the Ministry of Human Resource Development for fellowship to LT and SS.
Funding
Computational analyses were done on the Har Gobind Khorana Computational Biology cluster established and maintained by combining funds from IISER Bhopal under Grant # INST/BIO/2017/019, IYBA 2018 from the Department of Biotechnology (Grant no. BT/11/IYBA/2018/03), and ECRA from Science and Engineering Research Board (Grant no. ECR/2017/001430).
Author information
Authors and Affiliations
Contributions
LT and SS wrote the manuscript with NV. LT and SS analyzed the data and generated the results. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Teekas, L., Sharma, S. & Vijay, N. Lineage-specific protein repeat expansions and contractions reveal malleable regions of immune genes. Genes Immun 23, 218–234 (2022). https://doi.org/10.1038/s41435-022-00186-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41435-022-00186-4