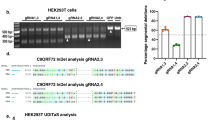
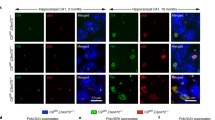
Abstract
Amyotrophic lateral sclerosis (ALS) is a fatal neurodegenerative disease that affects motor neurons, causing progressive muscle weakness and respiratory failure. The presence of an expanded hexanucleotide repeat in chromosome 9 open reading frame 72 (C9ORF72) is the most frequent mutation causing familial ALS and frontotemporal dementia (FTD). To determine if suppressing expression of C9ORF72 gene products can reduce toxicity, we designed a set of artificial microRNAs (amiRNA) targeting the human C9ORF72 gene. Here we report that an AAV9-mediated amiRNA significantly suppresses expression of the C9ORF72 mRNA, protein, and toxic dipeptide repeat proteins generated by the expanded repeat in the brain and spinal cord of C9ORF72 transgenic mice.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Chiò A, Logroscino G, Traynor BJ, Collins J, Simeone JC, Goldstein LA, et al. Global epidemiology of amyotrophic lateral sclerosis: a systematic review of the published literature. Neuroepidemiology. 2013;41:118–30.
Wijesekera LC, Leigh PN. Amyotrophic lateral sclerosis. Orphanet J Rare Dis. 2009;4:3.
DeJesus-Hernandez M, Mackenzie IRR, Boeve BFF, Boxer ALL, Baker M, Rutherford NJJ, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-Linked FTD and ALS. Neuron. 2011;72:245–56.
Majounie E, Renton AE, Mok K, Dopper EGPP, Waite A, Rollinson S, et al. Frequency of the C9orf72 hexanucleotide repeat expansion in patients with amyotrophic lateral sclerosis and frontotemporal dementia: a cross-sectional study. Lancet Neurol. 2012;11:323–30.
Renton AE, Majounie E, Waite A, Simón-Sánchez J, Rollinson S, Gibbs JR, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–68.
Devenney E, Hornberger M, Irish M, Mioshi E, Burrell J, Tan R, et al. Frontotemporal dementia associated with the C9ORF72 mutation: a unique clinical profile. JAMA Neurol. 2014;71:331–9.
Van Mossevelde S, van der Zee J, Cruts M, Van Broeckhoven C. Relationship between C9orf72 repeat size and clinical phenotype. Curr Opin Genet Dev. 2017;44:117–24.
Iacoangeli A, Al Khleifat A, Jones AR, Sproviero W, Shatunov A, Opie-Martin S, et al. C9orf72 intermediate expansions of 24–30 repeats are associated with ALS. Acta Neuropathologica. Communications. 2019;7:115.
Amick J, Ferguson SM. C9orf72: At the intersection of lysosome cell biology and neurodegenerative disease. Traffic. 2017;18:267–76.
Shi Y, Lin S, Staats KA, Li Y, Chang WH, Hung ST, et al. Haploinsufficiency leads to neurodegeneration in C9ORF72 ALS/FTD human induced motor neurons. Nat Med. 2018;24:313–25.
Zhu Q, Jiang J, Gendron TF, McAlonis-Downes M, Jiang L, Taylor A, et al. Reduced C9ORF72 function exacerbates gain of toxicity from ALS/FTD-causing repeat expansion in C9orf72. Nat Neurosci. 2020;23:615–24.
Mizielinska S, Lashley T, Norona FE, Clayton EL, Ridler CE, Fratta P, et al. C9orf72 frontotemporal lobar degeneration is characterised by frequent neuronal sense and antisense RNA foci. Acta Neuropathol. 2013;126:845–57.
Gendron TF, Bieniek KF, Zhang YJJ, Jansen-West K, Ash PEAA, Caulfield T, et al. Antisense transcripts of the expanded C9ORF72 hexanucleotide repeat form nuclear RNA foci and undergo repeat-associated non-ATG translation in c9FTD/ALS. Acta Neuropathol. 2013;126:829–44.
Mori K, Weng SM, Arzberger T, May S, Rentzsch K, Kremmer E, et al. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science. 2013;339:1335–8.
Zu T, Liu Y, Banez-Coronel M, Reid T, Pletnikova O, Lewis J, et al. RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc Natl Acad Sci USA. 2013;110:E4968–77.
Ash PEA, Bieniek KF, Gendron TF, Caulfield T, Lin WL, DeJesus-Hernandez M, et al. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron. 2013;77:639–46.
May S, Hornburg D, Schludi MH, Arzberger T, Rentzsch K, Schwenk BM, et al. C9orf72 FTLD/ALS-associated Gly-Ala dipeptide repeat proteins cause neuronal toxicity and Unc119 sequestration. Acta Neuropathol. 2014;128:485–503.
Wen X, Tan W, Westergard T, Krishnamurthy K, Markandaiah SS, Shi Y, et al. Antisense proline-arginine RAN dipeptides linked to C9ORF72-ALS/FTD form toxic nuclear aggregates that initiate in vitro and in vivo neuronal death. Neuron. 2014;84:1213–25.
Sun Y, Eshov A, Zhou J, Isiktas AU, Guo JU. C9orf72 arginine-rich dipeptide repeats inhibit UPF1-mediated RNA decay via translational repression. Nat Commun. 2020;11:3354.
West RJH, Sharpe JL, Voelzmann A, Munro AL, Hahn I, Baines RA, et al. Co-expression of C9orf72 related dipeptide-repeats over 1000 repeat units reveals age- and combination-specific phenotypic profiles in Drosophila. Acta Neuropathol Commun. 2020;8:158.
Zhang YJ, Guo L, Gonzales PK, Gendron TF, Wu Y, Jansen-West K, et al. Heterochromatin anomalies and double-stranded RNA accumulation underlie C9orf72 poly(PR) toxicity. Science. 2019;363:eaav2606.
Zhang YJ, Gendron TF, Ebbert MTW, O’Raw AD, Yue M, Jansen-West K, et al. Poly(GR) impairs protein translation and stress granule dynamics in C9orf72-associated frontotemporal dementia and amyotrophic lateral sclerosis. Nat Med. 2018;24:1136–42.
Miller RG, Mitchell JD, Moore DH. Riluzole for amyotrophic lateral sclerosis (ALS)/motor neuron disease (MND). Cochrane Database Syst Rev. 2012;2012:CD001447.
Takei K, Takahashi F, Liu S, Tsuda K, Palumbo J. Post-hoc analysis of randomised, placebo-controlled, double-blind study (MCI186-19) of edaravone (MCI-186) in amyotrophic lateral sclerosis. Amyotroph Lateral Scler Frontotemporal Degener. 2017;18:49–54.
Cruz MP. Edaravone (Radicava): a novel neuroprotective agent for the treatment of amyotrophic lateral sclerosis. P T. 2018;43:25–8.
Aschenbrenner DS. New drug approved for ALS. Am J Nurs. 2023;123:22.
O’Rourke JG, Bogdanik L, Muhammad AKMG, Gendron TF, Kim KJ, Austin A, et al. C9orf72 BAC transgenic mice display typical pathologic features of ALS/FTD. Neuron. 2015;88:892–901.
Schoch KM, Miller TM. Antisense oligonucleotides: translation from mouse models to human neurodegenerative diseases. Neuron. 2017;94:1056–70.
Borel F, Gernoux G, Sun H, Stock R, Blackwood M, Brown RH, et al. Safe and effective superoxide dismutase 1 silencing using artificial microRNA in macaques. Sci Transl Med. 2018;10:eaau6414.
Bak M, Silahtaroglu A, Møller M, Christensen M, Rath MF, Skryabin B, et al. MicroRNA expression in the adult mouse central nervous system. RNA. 2008;14:432–44.
Adlakha YK, Saini N. Brain microRNAs and insights into biological functions and therapeutic potential of brain enriched miRNA-128. Mol Cancer. 2014;13:33.
Peters OM, Cabrera GT, Tran H, Gendron TF, McKeon JE, Metterville J, et al. Human C9ORF72 hexanucleotide expansion reproduces RNA foci and dipeptide repeat proteins but not neurodegeneration in BAC transgenic mice. Neuron. 2015;88:902–9.
Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97.
Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–98.
Ambros V. The evolution of our thinking about microRNAs. Nat Med. 2008;14:1036–40.
Hoffer P, Ivashuta S, Pontes O, Vitins A, Pikaard C, Mroczka A, et al. Posttranscriptional gene silencing in nuclei. Proc Natl Acad Sci USA. 2011;108:409–14.
Huang V, Li LC. miRNA goes nuclear. RNA Biol. 2012;9:269–73.
Nishi K, Nishi A, Nagasawa T, Ui-Tei K. Human TNRC6A is an Argonaute-navigator protein for microRNA-mediated gene silencing in the nucleus. RNA. 2013;19:17–35.
Martier R, Liefhebber JM, Miniarikova J, van der Zon T, Snapper J, Kolder I, et al. Artificial microRNAs targeting C9orf72 can reduce accumulation of intra-nuclear transcripts in ALS and FTD patients. Mol Ther Nucleic Acids. 2019;14:593–608.
Batra R, Lee CW. Mouse models of C9orf72 hexanucleotide repeat expansion in amyotrophic lateral sclerosis/ frontotemporal dementia. Front Cell Neurosci. 2017;11:196.
Jiang J, Zhu Q, Gendron TF, Saberi S, McAlonis-Downes M, Seelman A, et al. Gain of toxicity from ALS/FTD-linked repeat expansions in C9ORF72 is alleviated by antisense oligonucleotides targeting GGGGCC-containing RNAs. Neuron. 2016;90:535–50.
Liu Y, Pattamatta A, Zu T, Reid T, Bardhi O, Borchelt DR, et al. C9orf72 BAC mouse model with motor deficits and neurodegenerative features of ALS/FTD. Neuron. 2016;90:521–34.
Burberry A, Suzuki N, Wang JY, Moccia R, Mordes DA, Stewart MH, et al. Loss-of-function mutations in the C9ORF72 mouse ortholog cause fatal autoimmune disease. Sci Transl Med. 2016;8:347ra93.
McLean JR, Smith GA, Rocha EM, Hayes MA, Beagan JA, Hallett PJ, et al. Widespread neuron-specific transgene expression in brain and spinal cord following synapsin promoter-driven AAV9 neonatal intracerebroventricular injection. Neurosci Lett. 2014;576:73–8.
Aschauer DF, Kreuz S, Rumpel S. Analysis of transduction efficiency, tropism and axonal transport of AAV serotypes 1, 2, 5, 6, 8 and 9 in the mouse brain. PLoS ONE. 2013;8:e76310.
Foust KD, Nurre E, Montgomery CL, Hernandez A, Chan CM, Kaspar BK. Intravascular AAV9 preferentially targets neonatal neurons and adult astrocytes. Nat Biotechnol. 2009;27:59–65.
Borel F, Gernoux G, Cardozo B, Metterville JP, Toro Cabreja GC, Song L, et al. Therapeutic rAAVrh10 mediated SOD1 silencing in adult SOD1G93A mice and nonhuman primates. Human Gene Therapy. 2016;27:19–31.
Pfister EL, Chase KO, Sun H, Kennington LA, Conroy F, Johnson E, et al. Safe and efficient silencing with a Pol II, but not a Pol lII, promoter expressing an artificial miRNA targeting human huntingtin. Mol Ther Nucleic Acids. 2017;7:324–34.
Pfister EL, DiNardo N, Mondo E, Borel F, Conroy F, Fraser C, et al. Artificial miRNAs reduce human mutant huntingtin throughout the striatum in a transgenic sheep model of Huntington’s disease. Hum Gene Ther. 2018;29:663–73.
Mueller C, Berry JD, McKenna-Yasek DM, Gernoux G, Owegi MA, Pothier LM, et al. SOD1 suppression with adeno-associated virus and microRNA in familial ALS. N Engl J Med. 2020;383:151–8.
Boeynaems S, Bogaert E, Michiels E, Gijselinck I, Sieben A, Jovičić A, et al. Drosophila screen connects nuclear transport genes to DPR pathology in c9ALS/FTD. Sci Rep. 2016;6:20877.
Sznajder ŁJ, Thomas JD, Carrell EM, Reid T, McFarland KN, Cleary JD, et al. Intron retention induced by microsatellite expansions as a disease biomarker. Proc Natl Acad Sci USA. 2018;115:4234–9.
Niblock M, Smith BN, Lee YB, Sardone V, Topp S, Troakes C, et al. Retention of hexanucleotide repeat-containing intron in C9orf72 mRNA: implications for the pathogenesis of ALS/FTD. Acta Neuropathol Commun. 2016;4:18.
Tabet R, Schaeffer L, Freyermuth F, Jambeau M, Workman M, Lee CZZ, et al. CUG initiation and frameshifting enable production of dipeptide repeat proteins from ALS/FTD C9ORF72 transcripts. Nat Commun. 2018;9:152–152.
Hautbergue GM, Castelli LM, Ferraiuolo L, Sanchez-Martinez A, Cooper-Knock J, Higginbottom A, et al. SRSF1-dependent nuclear export inhibition of C9ORF72 repeat transcripts prevents neurodegeneration and associated motor deficits. Nat Commun. 2017;8:16063.
Martier R, Liefhebber JM, García-Osta A, Miniarikova J, Cuadrado-Tejedor M, Espelosin M, et al. Targeting RNA-mediated toxicity in C9orf72 ALS and/or FTD by RNAi-based gene therapy. Mol Ther Nucleic Acids. 2019;16:26–37.
Liu Y, Andreucci A, Iwamoto N, Yin Y, Yang H, Liu F, et al. WVE-004, an investigational stereopure antisense oligonucleotide for the treatment of amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) (2302). Neurology. 2021;96. https://n.neurology.org/content/96/15_Supplement/2302
Tran H, Moazami MP, Yang H, McKenna-Yasek D, Douthwright CL, Pinto C, et al. Suppression of mutant C9orf72 expression by a potent mixed backbone antisense oligonucleotide. Nat Med. 2022;28:117–24.
Meijboom KE, Brown RH. Approaches to gene modulation therapy for ALS. Neurotherapeutics. 2022;19:1159–79.
Wang D, Tai PWL, Gao G. Adeno-associated virus vector as a platform for gene therapy delivery. Nat Rev Drug Discov. 2019;18:358–78.
Acknowledgements
We acknowledge G. Gao and Q. Su at University of Massachusetts Medical School who supervised the vector production. We thank Lorelei Stoica for training on all mouse surgeries, Lina Song for aiding with plasmid cloning and instruction, Qiushi Tang for assisting with vector constructs, Meghan Blackwood for aiding with tissue collections, Ozgun Uyan for scientific conversations and suggestions, and Allison Keeler for scientific conversations and editing the manuscript.
Funding
This study was supported by ALS Alliance Therapy (RHB), the ALS-FindingACure (RHB), and NINDS NS088689 (C.M. and RHB). RHB also received funding from the NINDS (NS079836), the ALS Association, ALSOne, the Angel Fund, the Cellucci Fund for ALS Research, the Michael Rosenfeld ALS Fund, the Pierre L. de Bourgknecht ALS Research Foundation, Project ALS and Target ALS.
Author information
Authors and Affiliations
Contributions
GTC led design, analysis, interpretation of the study, and prepared the manuscript. Performed screenings, cultures, surgeries, ddPCR analysis, RNA FlSH and quantifications. KEM prepared manuscript for publication, did analyses and statistics and performed immunohistochemistry and microscopy experiments. AA aided with surgeries and temporal injections, primary cultures, scientific input, and editing. HT performed MSD ELISA assays for DPR detection, aided with scientific input. ZF performed animal tissue collection and sectioning including RNAscope quantification. AW managed the mice colony and aided with stereotaxic surgeries. NW performed Western blots for detection of C9ORF72 protein. RS performed cryosections of spinal cord and LCM microdissections. TG performed ELISA assays for DPR detection. AGr aided with temporal injections and manuscript editing. AGi aided with primary culture dissection. RHB and CM conceived of the project and supervised all aspects of its execution and analysis.
Corresponding authors
Ethics declarations
Competing interests
RHB and CM are inventors on the patent for the technology described within this report (rAAV-Based Compositions and Methods for Treating Amyotrophic Lateral Sclerosis, WO2015143078A1) and may be entitled to royalty payments in the future. The patent has been licensed to Apic-Bio for which CM and RHB are founders with equity. All other authors declare no competing financial interests.
Ethical approvals
All mouse experiments were conducted at UMass Medical School following protocols approved by the Institutional Review Board. The University of Massachusetts Medical School Institutional Animal Care and Use Committee approved all experiments involving animals.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Cabrera, G.T., Meijboom, K.E., Abdallah, A. et al. Artificial microRNA suppresses C9ORF72 variants and decreases toxic dipeptide repeat proteins in vivo. Gene Ther 31, 105–118 (2024). https://doi.org/10.1038/s41434-023-00418-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41434-023-00418-w