Abstract
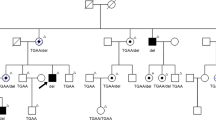
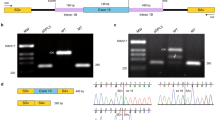
X-linked retinitis pigmentosa (XLRP) is the most severe form of Retinitis Pigmentosa (RP) and one of the leading causes of blindness in the world. Currently, there is no effective treatment for RP. In the present study, we recruited a XLRP family and identified a 4 bp deletion mutation (c. 2234_2237del) in RPGR ORF15 with Sanger sequencing, which was located in the exact same region as the missing XES (X chromosome exome sequencing) coverage. Then, we generated cell lines harboring the identified mutation and corrected it via enhanced prime editing system (ePE). Collectively, Sanger sequencing identified a pathogenic mutation in RPGR ORF15 for XLRP which was corrected with ePE. This study provides a valuable insight for genetic counseling of the afflicted family members and prenatal diagnosis, also paves a way for applying prime editing based gene therapy in those patients.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
The corresponding author declares that the data are available if requested. The deep sequencing data are available at the NCBI Sequence Read Archive (SRA) under BioProject PRJNA#573504 and PRJNA783078 (SRA accession number SRR17023438 and SRP17023439, sample accession numbers, SAMN23416124).
References
Verbakel SK, van Huet RAC, Boon CJF, den Hollander AI, Collin RWJ, Klaver CCW, et al. Non-syndromic retinitis pigmentosa. Prog Retin Eye Res. 2018;66:157–86.
Hartong DT, Berson EL, Dryja TP. Retinitis pigmentosa. Lancet. 2006;368:1795–809.
Dias MF, Joo K, Kemp JA, Fialho SL, Cunha AdS,Jr., Woo SJ. et al. Molecular genetics and emerging therapies for retinitis pigmentosa: Basic research and clinical perspectives. Prog Retin Eye Res. 2018;63:107–31.
Anasagasti A, Irigoyen C, Barandika O, Lopez de Munain A, Ruiz-Ederra J. Current mutation discovery approaches in retinitis pigmentosa. Vision Res. 2012;75:117–29.
Nash BM, Wright DC, Grigg JR, Bennetts B, Jamieson RV. Retinal dystrophies, genomic applications in diagnosis and prospects for therapy. Transl Pediatr. 2015;4:139–63.
Bassuk AG, Sujirakul T, Tsang SH, Mahajan VB. A novel RPGR mutation masquerading as Stargardt disease. Brit J Ophthalmol. 2014;98:709–11.
Berger W, Kloeckener-Gruissem B, Neidhardt J. The molecular basis of human retinal and vitreoretinal diseases. Prog Retin Eye Res. 2010;29:335–75.
Daiger SP, Sullivan LS, Bowne SJ. Genes and mutations causing retinitis pigmentosa. Clin Genet. 2013;84:132–41.
Zhao L, Wang F, Wang H, Li Y, Alexander S, Wang K, et al. Next-generation sequencing-based molecular diagnosis of 82 retinitis pigmentosa probands from Northern Ireland. Hum Genet. 2015;134:217–30.
Ferrari S, Di Iorio E, Barbaro V, Ponzin D, Sorrentino FS, Parmeggiani F. Retinitis pigmentosa: genes and disease mechanisms. Curr Genomics. 2011;12:238–49.
Bamshad MJ, Ng SB, Bigham AW, Tabor HK, Emond MJ, Nickerson DA, et al. Exome sequencing as a tool for Mendelian disease gene discovery. Nat Rev Genet. 2011;12:745–55.
Koboldt DC, Steinberg KM, Larson DE, Wilson RK, Mardis ER. The next-generation sequencing revolution and its impact on genomics. Cell. 2013;155:27–38.
Giacalone JC, Andorf JL, Zhang Q, Burnight ER, Ochoa D, Reutzel AJ, et al. Development of a molecularly stable gene therapy vector for the treatment of RPGR-associated X-linked retinitis pigmentosa. Hum Gene Ther. 2019;30:967–74.
Diakatou M, Manes G, Bocquet B, Meunier I, Kalatzis V Genome editing as a treatment for the most prevalent causative genes of autosomal dominant retinitis pigmentosa. Int J Mol Sci. 2019;20.
Collin C, Liu Q CRISPR/Cas9-based genome editing approaches for RP1 associated autosomal dominant Retinitis Pigmentosa. Invest Ophthalmol Vis Sci. 2021;62.
Quinn J, Musa A, Kantor A, McClements ME, Cehajic-Kapetanovic J, MacLaren RE, et al. Genome-editing strategies for treating human retinal degenerations. Hum Gene Ther. 2021;32:247–59.
Wu WH, Tsai YT, Justus S, Lee TT, Zhang L, Lin CS, et al. CRISPR repair reveals causative mutation in a preclinical model of retinitis pigmentosa. Mol Ther. 2016;24:1388–94.
Bakondi B, Lv W, Lu B, Jones MK, Tsai Y, Kim KJ, et al. In vivo CRISPR/Cas9 gene editing corrects retinal dystrophy in the S334ter-3 rat model of autosomal dominant retinitis pigmentosa. Mol Ther. 2016;24:556–63.
Hu S, Du J, Chen NN, Jia RX, Zhang JL, Liu XZ, et al. In vivo CRISPR/Cas9-mediated genome editing mitigates photoreceptor degeneration in a mouse model of X-linked retinitis pigmentosa. Invest Ophthalmol Vis Sci. 2020;61:10.
Bassuk AG, Zheng A, Li Y, Tsang SH, Mahajan VB. Precision medicine: genetic repair of retinitis pigmentosa in patient-derived stem cells. Sci Rep. 2016;6:1–6.
Ihry RJ, Worringer KA, Salick MR, Frias E, Ho D, Theriault K, et al. p53 inhibits CRISPR-Cas9 engineering in human pluripotent stem cells. Nat Med. 2018;24:939–46.
Anzalone AV, Koblan LW, Liu DR. Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors. Nat Biotechnol. 2020;38:824–44.
Anzalone AV, Randolph PB, Davis JR, Sousa AA, Koblan LW, Levy JM, et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature. 2019;576:149–57.
Liu Y, Yang G, Huang S, Li X, Wang X, Li G, et al. Enhancing prime editing by Csy4-mediated processing of pegRNA. Cell Res. 2021;31:1134–6.
Wu J, Shen E, Shi D, Sun Z, Cai T. Identification of a novel Cys146X mutation of SOD1 in familial amyotrophic lateral sclerosis by whole-exome sequencing. Genet Med. 2012;14:823–6.
Wang T, Liu Q, Li X, Wang X, Li J, Zhu X, et al. RRBS-analyser: a comprehensive web server for reduced representation bisulfite sequencing data analysis. Hum Mutat. 2013;34:1606–10.
Han R, Wang X, Wang D, Wang L, Yuan Z, Ying M, et al. GPR143 Gene Mutations in Five Chinese Families with X-linked Congenital Nystagmus. Sci Rep. 2015;5:12031.
Lv X, Qiu K, Tu T, He X, Peng Y, Ye J, et al. Development of a simple and quick method to assess base editing in human cells. Mol Ther-Nucl Acids. 2020;20:580–8.
Wang Y, Wang B, Xie H, Ren Q, Liu X, Li F, et al. Efficient human genome editing using SaCas9 ribonucleoprotein complexes. Biotechnol J. 2019;14:e1800689.
Zhang X, Ge X, Yu Y, Zhang Y, Wu Y, Luan Y, et al. Identification of three novel mutations in the FRMD7 gene for X-linked idiopathic congenital nystagmus. Sci Rep. 2014;4:3745.
Vervoort R, Lennon A, Bird AC, Tulloch B, Axton R, Miano MG, et al. Mutational hot spot within a new RPGR exon in X-linked retinitis pigmentosa. Nat Genet. 2000;25:462–6.
Breuer DK, Yashar BM, Filippova E, Hiriyanna S, Lyons RH, Mears AJ, et al. A comprehensive mutation analysis of RP2 and RPGR in a north American cohort of families with X-linked retinitis pigmentosa. Am J Hum Genet. 2002;70:1545–54.
Kim C, Kim KJ, Bok J, Lee E-J, Kim D-J, Oh JH, et al. Microarray-based mutation detection and phenotypic characterization in Korean patients with retinitis pigmentosa. Mol Vis. 2012;18:2398–410.
Blanco-Kelly F, García-Hoyos M, Cortón M, Avila-Fernández A, Riveiro-Álvarez R, Giménez A, et al. Genotyping microarray: mutation screening in Spanish families with autosomal dominant retinitis pigmentosa. Mol Vis. 2012;18:1478–83.
Zhong YM, Xu F, Wu JH, Schubert J, Li MM. Application of next generation sequencing in laboratory medicine. Ann Lab Med. 2021;41:25–43.
Jennings LJ, Arcila ME, Corless C, Kamel-Reid S, Lubin IM, Pfeifer J, et al. Guidelines for validation of next-generation sequencing-based oncology panels a joint consensus recommendation of the association for molecular pathology and college of american pathologists. J Mol Diagn. 2017;19:341–65.
Cromer MK, Camarena J, Martin RM, Lesch BJ, Vakulskas CA, Bode NM, et al. Gene replacement of α-globin with β-globin restores hemoglobin balance in β-thalassemia-derived hematopoietic stem and progenitor cells. Nat Med. 2021;27:677–87.
Guha TK, Wai A, Hausner G. Programmable genome editing tools and their regulation for efficient genome engineering. Comput Struct Biotec. 2017;15:146–60.
Jiang T, Zhang X-O, Weng Z, Xue W. Deletion and replacement of long genomic sequences using prime editing. Nat Biotechnol. 2022;40:227–34.
Choi J, Chen W, Suiter CC, Lee C, Chardon FM, Yang W, et al. Precise genomic deletions using paired prime editing. Nat Biotechnol. 2022;40:218–26.
Chen PJ, Hussmann JA, Yan J, Knipping F, Ravisankar P, Chen P-F, et al. Enhanced prime editing systems by manipulating cellular determinants of editing outcomes. Cell. 2021;184:5635–52.
Zhuang Y, Liu J, Wu H, Zhu Q, Yan Y, Meng H, et al. Increasing the efficiency and precision of prime editing with guide RNA pairs. Nat Chem Biol. 2022;18:29–37.
Acknowledgements
We thank the families for their participation in this project. We are indebted to Dr. Jinyu Wu (Institute of Genomic Medicine, Wenzhou Medical University) for bioinformatics analysis and Prof.Peter Reinach for comments on the manuscript. This work was supported by grants from National Natural Science Foundation of China (81201181), Science Technology project of Zhejiang Province (2017C37176), and Project of State Key Laboratory of Ophthalmology, Optometry and Visual Science, Wenzhou Medical University (J02–20190201) and Wenzhou City Grant (H20210011).
Author information
Authors and Affiliations
Contributions
XL and FG conceived and designed the study experiments. ZZ and XZ identified the mutation. XL, ZZ, DX, and FG wrote and edited manuscript. XL, YZ, JL, YZ, BW, SL, WS performed the experiments. ZS, JX, JQ, and FG contributed to data analysis, data interpretation. XL, DX, and FG revised the manuscript. All authors have read and approved the final manuscript, and therefore, have full access to all data in this study and take responsibility for the integrity and security of the data.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1975, as revised in 2008 (5). Informed consent was obtained from all patients for being included in the study. Additional informed consent was obtained from all patients for which identifying information is included in this article.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Lv, X., Zheng, Z., Zhi, X. et al. Identification of RPGR ORF15 mutation for X-linked retinitis pigmentosa in a large Chinese family and in vitro correction with prime editor. Gene Ther 30, 160–166 (2023). https://doi.org/10.1038/s41434-022-00352-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41434-022-00352-3
This article is cited by
-
X-Linked Retinitis Pigmentosa Gene Therapy: Preclinical Aspects
Ophthalmology and Therapy (2023)