Abstract
Finnish gelsolin amyloidosis (AGel amyloidosis) is an autosomal dominantly inherited systemic disorder with ophthalmologic, neurologic and dermatologic symptoms. Only the gelsolin (GSN) c.640G>A variant has been found in the Finnish patients thus far. The purpose of this study was to examine whether the Finnish patients have a common ancestor or whether multiple mutation events have occurred at c.640G, which is a known mutational hot spot. A total of 79 Finnish AGel amyloidosis families including 707 patients were first discovered by means of patient interviews, genealogic studies and civil and parish registers. From each family 1-2 index patients were chosen. Blood samples were available from 71 index patients representing 64 families. After quality control, SNP array genotype data were available from 68 patients from 62 nuclear families. All the index patients had the same c.640G>A variant (rs121909715). Genotyping was performed using the Illumina CoreExome SNP array. The homozygosity haplotype method was used to analyse shared haplotypes. Haplotype analysis identified a shared haplotype, common to all studied patients. This shared haplotype included 17 markers and was 361 kb in length (GRCh37 coordinates 9:124003326–124364349) and this level of haplotype sharing was found to occur highly unlikely by chance. This GSN haplotype ranked as the largest shared haplotype in the 68 patients in a genome-wide analysis of haplotype block lengths. These results provide strong evidence that although there is a known mutational hot spot at GSN c.640G, all of the studied 62 Finnish AGel amyloidosis families are genetically linked to a common ancestor.
Similar content being viewed by others
Introduction
Finnish type of hereditary gelsolin amyloidosis (AGel amyloidosis) (OMIM (Online Mendelian Inheritance in Man) #105120) or Meretoja’s disease is an autosomal dominant systemic amyloidosis [1]. The number of patients in Finland is estimated between 400 and 1000 [2, 3], which makes it one of the most common diseases of the Finnish disease heritage [4]. AGel amyloidosis has also been found in many other European countries as well as in North America [5,6,7], South America [8,9,10] and Asia [11, 12], [13, 14]. The Finnish variant and most other ethnic variants are caused by a mutation in the gelsolin (GSN) gene, located on chromosome 9 at q33.2 [15], where guanine is replaced by adenosine (NM_000177.4:c.640G>A, NP_000168.1:p.Asp214Asn) [16, 17]. A less common variant in the same locus c.640G>T has been reported to cause AGel amyloidosis in Denmark, former Czechoslovakia, France, Brazil [5], South Korea [14] and the United States [7]. In addition to this, two recently identified GSN variants (c.633C>A and c.580G>A) have been described to cause renal amyloidosis in North America [18, 19].
The symptoms of AGel amyloidosis are thought to be a consequence of generation and deposition of GSN amyloid (AGel) fibrils and pre-amyloid oligomers, which originate from the misfolded variant GSN molecule and accumulate in tissues [20]. These symptoms include particularly ophthalmologic, neurologic and dermatologic signs [5]. These signs are recently demonstrated to progress with increasing AGel deposition [21]. The penetrance of the GSN c.640G>A variant is 100%, but the manifestations of the disease may vary significantly between different patients [5]. For the moment, patients can only be offered symptomatic treatment. However prominent amyloid elastosis in dermal [22] and vascular tissues [23] may represent an important target for future therapeutic strategies with anti-amyloidogenic molecules, such as, e.g., AGel-specific nanobodies [24,25,26].
All the Finnish patients examined so far carry the same GSN c.640G>A variant. It is not known whether this is due to a mutational hot spot in the gelsolin gene, which has been the prevailing hypothesis concerning the gene variants in different ethnic groups [27, 28], or due to a common ancestor. Dr. Meretoja, a Finnish ophthalmologist who first described the disease in 1969 [1], suggested that all the Finnish patients with this amyloidosis originate from one common ancestor who lived over 600 years ago [3]. This hypothesis has been later strengthened by the demonstration of a homogeneous GSN c.640G>A variant [29] and the uniformity of the disease associated haplotypes in 10 and 7 studied Finnish families [27, 28]. However, a more comprehensive study on the subject has not been available. We first identified 79 Finnish AGel amyloidosis families using methods of classical genealogy. After that we conducted a haplotype analysis study. This included at least one index patient from all available Finnish AGel amyloidosis families. This study was conducted in order to find out whether the Finnish patients share a common haplotype, indicative of a common ancestor.
Materials and methods
Genealogic study
A national registry for AGel amyloidosis patients in Finland, FIN-GAR (The Finnish Gelsolin Amyloidosis Patient Registry), was founded in 2013 [30]. In the process, information on family history including affected individuals and places of domiciles were collected by a questionnaire. Majority (87% of the patients in the registry) were also interviewed over telephone in order to get more detailed data. In addition to this, some patients were able to give written genealogy studies about their family. Everything mentioned above was combined with already existing AGel amyloidosis family data since the end of 1980s from previous studies of our research group. Access to the civil registry and parish registers was also used in order to find information about the ancestors of the patients. Using classical genealogy methods AGel amyloidosis families were traced down as far as possible.
Subject selection for haplotype analysis
Because all family members of the same family affected with AGel amyloidosis were assumed to share the same disease haplotype, we invited one or two index patients from each identified family to participate in the haplotype analysis study. All the invited patients had characteristic clinical manifestations of the disease and their diagnosis had been made by an ophthalmologist or other physician. In 38 patients the diagnosis had also been confirmed by a gene test which verified the variant c.640G>A in the gelsolin gene.
Only families including at least one member in FIN-GAR patient register (72 out of 79 families) were enroled in this study as they had given their consent to contact them for research purposes. After giving the informed consent, blood samples were collected from 71 index patients representing 64 families. After the quality control, samples from 68 index patients representing 62 families were left for analysis. The subject selection process is presented in Fig. 1.
Genotyping and haplotype analysis
Using the standard procedure of Wizard® Genomic DNA Purification kit the DNA was isolated from red blood cells. First, red blood cells and their nuclei were lysed, followed by protein precipitation. Finally, DNA was precipitated and rehydrated. Concentrations and purity of DNA samples were measured and samples were titrated to a predetermined concentration (50 ng/μl).
Samples were then genotyped with CoreExome single-nucleotide polymorphism (SNP) arrays and GenomeStudio v. 2011.1 software (Illumina, Inc., San Diego, CA, USA). The SNP array contains different tag SNPs and detects these markers from samples. Additional quality control for identity by descent (IBD, to remove duplicate samples) and sex was performed using the PLINK software [31], which is a computer program for SNP data handling. GSN c.640G>A (rs121909715, GRCh37 chr9:g.124073097G>A) had been previously tested as part of diagnostics in most cases. We sequenced the gelsolin gene variant site in 33 patients by Sanger sequencing using the following primers for generating the amplicon: 5ʹ-CACTGTGTTGCTGGAAAGCC-3ʹ and 5ʹ-CGTGGATGTACCCAAGTCCC-3ʹ. The genotyping data are publically available at the figshare database (dx.doi.org/10.6084/m9.figshare.5455495).
Patients’ haplotypes were analysed using the homozygosity haplotype method [32]. Markers with a minor allele frequency of at least 10% within the genotyped patient samples and a genotyping rate of 95% or more were included in the analysis. The 10% minor allele frequency cut-off was chosen to exclude markers with low informativeness. The likelihood of a random finding at the GSN locus was evaluated by two sampling methods. The first evaluation consisted of analysing 10,000 random permutations of the SNP array marker labels and running an identical homozygosity haplotype analysis for each such permutation of the data. For the second method of evaluation, a sample of equal size to the study cohort was repeatedly drawn from a larger population-based cohort (Vantaa 85+, 10,000 random samplings) and then the homozygosity haplotype analysis was performed for each sample. The Vantaa 85+ study sample [33] is a Finnish population-based cohort comprising all subjects aged ⩾85 years from Vantaa, Finland as of 1 April 1991 (n = 601). Whole-genome sequencing has been performed in a subsample, which was selected based on the available DNA amount. Whole-genome sequencing was performed in 309 samples with at least 4 µg good-quality DNA left; of these, 300 yielded good-quality sequence data. The sequencing was performed in Broad Institute using PCR-free sample preparation with Illumina HiseqX chemistry, and we achieved a mean coverage of 27×. Only markers present in the CoreExome SNP array were drawn from the whole-genome sequence data for the random sampling analysis.
The local Ethics Committee of the Helsinki University Hospital has approved the study.
Results
Finnish AGel amyloidosis families
Altogether, 79 AGel amyloidosis families in Finland could be traced down. After this, ancestors could no longer be identified by classical genealogy methods. Family trees included 2 to 106 patients.
The earliest suspected patients who could be identified were born in the 1820s. A total of 29 (37%) families originated from the Kymenlaakso area, which represents only 3% of the Finnish population. Lammi, a municipality located in Southern Tavastia, represented a second geographical concentration with 10 (13%) families. Also, 9 (11%) families originated from areas southeast of Finland that had been ceded to the Soviet Union after the Second World War in 1944 (Fig. 2).
The identified 79 families included 707 patients, 235 of whom were members of the FIN-GAR registry diagnosed by a physician. The remaining patients were identified as AGel amyloidosis patients based on reports of relatives and earlier material collected by the research group. Therefore, the method of diagnosis for them was unknown to us.
Genotyping
A total of 71 blood samples from 64 families were available for genotyping. These represent 81% of all identified 79 Finnish AGel amyloidosis families. All 71 genotyped samples had a genotyping success rate of over 99%. Two samples were removed from the analysis due to high degree (100%) of IBD with each other and one sample was ruled out because of gender mismatch. After quality control, 68 patient samples representing 62 families were available for final analysis. These families represent 78% of all currently identified families. The baseline information of the patients from these 62 families is given in Table 1.
Founder haplotype analysis
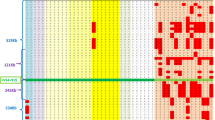
All patients had the GSN c.640G>A variant. We wanted to test if these patients share a common founder haplotype at the GSN locus. Using the homozygosity haplotype method we identified a shared 17-SNP haplotype at the GSN locus spanning 361 kb (GRCh37 coordinates 9:124003326–124364349) (Fig. 3). By permutating the marker data we estimated the likelihood of this finding by chance and found it highly unlikely (zero occurrence in 10,000 simulated permutations of the data). Moreover, in a genome-wide analysis of the SNP array data, this was the largest haplotype shared by all patients (Fig. 4).
Genome-wide shared haplotype analysis. Shared haplotypes were detected using the homozygosity haplotype method from SNP array data. The highest peak corresponding to the largest detected shared haplotype between all subjects encompasses the gelsolin gene area and contains the gelsolin (GSN) c.640G>A variant
Next, we tested whether the GSN locus haplotype sharing is a common feature of the Finnish population. We empirically tested shared haplotypes in a population-based sample of elderly Finns (Vantaa 85+, n = 300) using the same marker set and same methodology. The same number of samples (n = 68) was drawn randomly 10,000 times, and we calculated how many times GSN locus appeared as a shared haplotype. This analysis showed no shared haplotype at the GSN locus in any of the 10,000 random samplings. Furthermore, we estimated the population frequency of the 17-SNP haplotype in the full Vantaa 85+ data by analysing subjects carrying at least one copy of the alleles present in the haplotype. This procedure yielded a frequency of 0.143, which is the theoretical maximum haplotype frequency in the Vantaa 85+ sample, and it thus appears highly unlikely that 68 subjects would share this haplotype by chance. Taken together, these observations support the concept that all of the patients share a common ancestor.
Discussion
We present here the so far largest haplotype analysis in AGel amyloidosis, which provides strong evidence indicating that all the analysed 62 Finnish AGel amyloidosis families have a common ancestor.
It has been previously suggested that there is a mutational hot spot in this specific location c.640G [27, 28]. For this reason it is possible that sporadic GSN c.640G>A mutations have taken place independently in several persons in Finland. This has been proposed to cause AGel amyloidosis in different ethnic groups [27, 28]. However, our data indicate that in Finland this is not the case. On the contrary, the cases of AGel amyloidosis, reported in some families in Japan [28] and the Netherlands [27], seem to have arisen from sporadic de novo mutations and at least two independent haplotypes have been reported in Japan [34]. One family in the United States has been identified to carry the same haplotype as the Finnish AGel amyloidosis families [35]. However in most non-Finnish GSN c.640G>A families worldwide haplotype analyses have not been performed.
Among the 62 families, there were 61 who shared a very large haplotype of 1290 kb with 55 markers (coordinates 9:123074244–124364349). Table 2 shows the markers of both 361 kb and 1290 kb haplotypes. Thus, 98% of the families have a larger haplotype and a single family reduced the shared haplotype block to 361 kb. This indicates that a recent recombination event has taken place in this family between markers rs10818516 and rs10760160 (Table 2). Most of the families presented here originate from the Kymenlaakso region in South Eastern Finland. Additionally, a large number of families originate from the municipality of Lammi in Southern Tavastia, located in Southern Finland. Areal concentration of the patients' places of birth is evident also when regarding other diseases of the Finnish disease heritage [4]. There are two background factors contributing in the formation of the Finnish disease heritage. First, national-level isolation influenced by geography, language, culture and religion has kept the original gene pool homogeneous for generations. Second, the Finnish people have typically stayed in their home region until the end of the Second World War. This regional isolation together with population expansion and genetic drift has enabled the accumulation of different disease genes in certain loci [4].
It is not known whether the common ancestor of the Finnish AGel amyloidosis patients originated from the Kymenlaakso area or whether he/she moved there from some unknown location. In this study this fact remains still open for question, because so far we could not trace the families further with classical genealogical methods in our search of the common ancestor. Dr Meretoja suggested that the common ancestor was a man called Matti of Oravalae who might have lived in the fourteenth or fifteenth century in Lammi. He would have brought the disease to Valkeala later on, located in the Northern Kymenlaakso [3]. Interestingly, 9 families originated from the Karelia region, indicating a possibility that the Finnish c.640G>A variant could have originated from there.
Even though the study includes an index patient of the large majority (78%) of the known Finnish AGel amyloidosis families, its coverage is not 100%. Getting a complete coverage is not feasible as there still might be some unidentified families and not all of the patients want to take part in studies.
This study represents the most comprehensive study on the Finnish AGel amyloidosis haplotype so far and together with the previous studies [27, 28] it indicates that most, if not all, known Finnish AGel amyloidosis patients share a common ancestor. In the future work on the classical genealogy concerning Finnish AGel amyloidosis patients will be continued in order to find out where the common ancestor came from. Investigating the age of the GSN c.640G>A variant would further clarify the history and the origin of the disease. In the following haplotype studies it would be interesting to examine further how widely this haplotype has spread among non-Finnish AGel amyloidosis families, and how much the disease is caused by sporadic hot spot mutations worldwide.
References
Meretoja J, et al. Familial systemic paramyloidosis with lattice dystrophy of the cornea, progressive cranial neuropathy, skin changes and various internal symptoms. A previously unrecognized heritable syndrome. Ann Clin Res 1969;1(4):314–24
Kiuru S. Familial amyloidosis of the Finnish type (FAF). Acta Neurol Scand 1992;86(4):346–53.
Meretoja J. Suomalainen amyloiditauti (the Finnish amyloid disease representing our disease inheritage). J Finn Med Assoc 1976;31:2234–6.
Norio R. Suomineidon geenit. Keuruu, Finland: Otava, 2000.
Kiuru-Enari S, Haltia M. Hereditary gelsolin amyloidosis. In: Said G, Krarup C (eds). Peripheral Nerve Disorders, Volume 115: Handbook of Clinical Neurology. Amsterdam: Elsevier; 2013. p. 659–81.
Gonzalez-Rodriguez J, Ramirez-Miranda A, Hernandez-Da Mota SE, Zenteno JC. TGFBI, CHST6, and GSN gene analysis in Mexican patients with stromal corneal dystrophies. Graefes Arch Clin Exp Ophthalmol 2014;252(8):1267–72.
Caress JB, Johnson JO, Abramzon YA, Hawkins GA, Gibbs JR, Sullivan EA, et al. Exome sequencing establishes a gelsolin mutation as the cause of inherited bulbar‐onset neuropathy. Muscle Nerve 2016;56:1001–5.
Spuler S, Emslie‐Smith A, Engel AG. Amyloid myopathy: an underdiagnosed entity. Ann Neurol 1998;43(6):719–28.
Felix EPV, Jung LS, Carvalho GS, Oliveira ASB. Corneal lattic dystrophy type II: familial amyloid neuropathy type IV (gelsolin amyloidosis). Einstein 2008;6:505–6.
Solari HP, Ventura MP, Antecka E, Belfort Junior R, Burnier Jr MN. Danish type gelsolin-related amyloidosis in a Brazilian family: case reports. Arq Bras Oftalmol 2011;74(4):286–8.
Maramattom BV, Chickabasaviah YT. A new Indian family affected by gelsolin amyloidosis. Neurol India 2013;61(6):673–5.
Ardalan MR, Shoja MM, Kiuru-Enari S. Amyloidosis-related nephrotic syndrome due to a G654A gelsolin mutation: the first report from the Middle East. Nephrol Dial Transplant 2007;22(1):272–5.
Chardon J, Massie R Progressive facial diparesis caused by gelsolin mutation in a Sri Lankan family. AANEM 61st Annual Meeting; 29 October to 1 November 2014; Savannah, GA, USA, 2014.
Park K-J, Park J-H, Park J-H, Cho EB, Kim BJ, Kim J-W. The first Korean family with hereditary gelsolin amyloidosis caused by p. D214Y mutation in the GSN gene. Ann Lab Med 2016;36(3):259–62.
Kwiatkowski D, Westbrook C, Bruns G, Morton C. Localization of gelsolin proximal to ABL on chromosome 9. Am J Hum Genet 1988;42(4):565–72.
Maury C, Kere J, Tolvanen R, De la Chapelle A. Finnish hereditary amyloidosis is caused by a single nucleotide substitution in the gelsolin gene. FEBS Lett 1990;276(1):75–7.
Levy E, Haltia M, Fernandez-Madrid I, Koivunen O, Ghiso J, Prelli F, et al. Mutation in gelsolin gene in Finnish hereditary amyloidosis. J Exp Med 1990;172(6):1865–7.
Efebera YA, Sturm A, Baack EC, Hofmeister CC, Satoskar A, Nadasdy T, et al. Novel gelsolin variant as the cause of nephrotic syndrome and renal amyloidosis in a large kindred. Amyloid 2014;21(2):110–2.
Sethi S, Theis JD, Quint P, Maierhofer W, Kurtin PJ, Dogan A, et al. Renal amyloidosis associated with a novel sequence variant of gelsolin. Am J Kidney Dis 2013;61(1):161–6.
Solomon JP, Page LJ, Balch WE, Kelly JW. Gelsolin amyloidosis: genetics, biochemistry, pathology and possible strategies for therapeutic intervention. Crit Rev Biochem Mol Biol 2012;47(3):282–96.
Pihlamaa T, Suominen S, Kiuru-Enari S, Tanskanen M. Increasing amount of amyloid are associated with the severity of clinical features in hereditary gelsolin (AGel) amyloidosis. Amyloid 2016;23:1–9.
Kiuru-Enari S, Somer H, Seppäläinen A-M, Notkola I-L, Haltia M. Neuromuscular pathology in hereditary gelsolin amyloidosis. J Neuropathol Exp Neurol 2002;61(6):565–71.
Koskelainen S, Pihlamaa T, Suominen S, Zhao F, Salo T, Risteli J, et al. Gelsolin amyloid angiopathy causes severe disruption of the arterial wall. APMIS 2016;124(8):639–48.
Van Overbeke W, Verhelle A, Everaert I, Zwaenepoel O, Vandekerckhove J, Cuvelier C, et al. Chaperone nanobodies protect gelsolin against MT1-MMP degradation and alleviate amyloid burden in the gelsolin amyloidosis mouse model. Mol Ther 2014;22(10):1768–78.
Van Overbeke W, Wongsantichon J, Everaert I, Verhelle A, Zwaenepoel O, Loonchanta A, et al. An ER-directed gelsolin nanobody targets the first step in amyloid formation in a gelsolin amyloidosis mouse model. Hum Mol Genet 2015;24(9):2492–507.
Lahoutte T, et al. Non-invasive imaging of amyloid deposits in a mouse model of AGel using 99mTc-modified nanobodies and SPECT/CT. Mol Imaging Biol 2016;18(6):887–97
de la Chapelle A, Tolvanen R, Boysen G, Santavy J, Bleeker-Wagemakers L, Maury C, et al. Gelsolin–derived familial amyloidosis caused by asparagine or tyrosine substitution for aspartic acid at residue 187. Nat Genet 1992;2(2):157–60.
Paunio T, Sunada Y, Kiuru S, Makishita H, Ikeda SI, Weissenbach J, et al. Haplotype analysis in gelsoiin‐related amyloidosis reveals independent origin of identical mutation (G654A) of gelsolin in Finland and Japan. Hum Mutat 1995;6(1):60–5.
Kiuru S. Gelsolin-related familial amyloidosis, Finnish type (FAF), and its variants found worldwide. Amyloid 1998;5(1):55–66.
Nikoskinen T, Schmidt E-K, Strbian D, Kiuru-Enari S, Atula S. Natural course of Finnish gelsolin amyloidosis. Ann Med 2015;47(6):506–11.
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007;81(3):559–75.
Miyazawa H, Kato M, Awata T, Kohda M, Iwasa H, Koyama N, et al. Homozygosity haplotype allows a genomewide search for the autosomal segments shared among patients. Am J Hum Genet 2007;80(6):1090–102.
Tanskanen M, Mäkelä M, Notkola IL, Myllykangas L, Rastas S, Oinas M, et al. Population‐based analysis of pathological correlates of dementia in the oldest old. Ann Clin Transl Neurol 2017;4(3):154–65.
Taira M, Ishiura H, Mitsui J, Takahashi Y, Hayashi T, Shimizu J, et al. Clinical features and haplotype analysis of newly identified Japanese patients with gelsolin-related familial amyloidosis of Finnish type. Neurogenetics 2012;13(3):237–43.
Steiner RD, Evans JP, Paunio T, Uemichi T, Benson MD. Asp187Asn mutation of gelsolin in an American kindred with familial amyloidosis, Finnish type (FAP IV). Hum Genet 1995;95(3):327–30.
Acknowledgements
We thank our Finnish AGel amyloidosis patients and their families as well as the patient organization SAMY (www.suomenamyloidoosiyhdistys.fi) for their considerable support and cooperation during this study. We also thank the laboratory technician Lilja Jansson for her substantial support in the laboratory work. The Vantaa 85+ whole-genome sequencing was funded by Merck Inc., and Intramural Research Program of the NIH and we thank Drs David Stone (Merck Inc.) and Bryan Traynor (NIH/NIA) for their valuable contribution in obtaining these data. We are grateful to the Kymenlaakso Regional Fund of the Finnish Cultural Foundation, Kymin100 foundation, Helsinki University Hospital and to Finska Läkaresällskapet for their financial support. The funders had no role in the study design, data collection and analysis or preparation of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Additional information
T Mustonen and EK Schmidt contributed equally to this work.
Rights and permissions
About this article
Cite this article
Mustonen, T., Schmidt, EK., Valori, M. et al. Common origin of the gelsolin gene variant in 62 Finnish AGel amyloidosis families. Eur J Hum Genet 26, 117–123 (2018). https://doi.org/10.1038/s41431-017-0026-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41431-017-0026-x
This article is cited by
-
Hearing problems in patients with hereditary gelsolin amyloidosis
Orphanet Journal of Rare Diseases (2021)