Abstract
Background
Epigenetic studies have reported relationships between dietary nutrient intake and methylation levels. However, genetic variants that may affect DNA methylation (DNAm) pattern, called methylation quantitative loci (mQTL), are usually overlooked in these analyses. We investigated whether mQTL change the relationship between dietary nutrient intake and leukocyte DNAm levels with an example of estimated fatty acid intake and ATP-binding cassette transporter A1 (ABCA1).
Methods
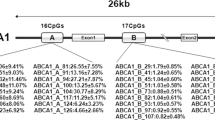
A cross-sectional study on 231 participants (108 men, mean age: 62.7 y) without clinical history of cancer and no prescriptions for dyslipidemia. We measured leukocyte DNAm levels of 8 CpG sites within ABCA1 gene by pyrosequencing method and used mean methylation levels for statistical analysis. TaqMan assay was used for genotyping a genetic variant of ABCA1 (rs1800976). Dietary fatty acid intake was estimated with a validated food frequency questionnaire and adjusted for total energy intake by using residual methods.
Results
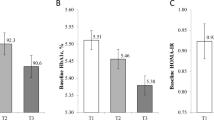
Mean ABCA1 DNAm levels were 5% lower with the number of minor alleles in rs1800976 (CC, 40.6%; CG, 35.9%; GG, 30.6%). Higher dietary n-3 PUFA intake was associated with lower ABCA1 DNAm levels (1st (ref) vs. 4th, β [95% CI]: –2.52 [–4.77, –0.28]). After controlling for rs180076, the association between dietary n-3 PUFA intake and ABCA1 DNAm levels was attenuated, but still showed an independent association (1st (ref) vs. 4th, β [95% CI]: –2.00 [–3.84, –0.18]). The interaction of mQTL and dietary n-3 PUFA intake on DNAm levels was not significant.
Conclusions
This result suggested that dietary n-3 PUFA intake would be an independent predictor of DNAm levels in ABCA1 gene after adjusting for individual genetic background. Considering mQTL need to broaden into other genes and nutrients for deeper understanding of DNA methylation, which can contribute to personalized nutritional intervention.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Data availability
The datasets generated during and/or analyzed during the current study are not publicly available but are available from the corresponding author on reasonable request.
References
Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21.
Boks MP, Derks EM, Weisenberger DJ, Strengman E, Janson E, Sommer IE, et al. The relationship of DNA methylation with age, gender and genotype in twins and healthy controls. PLoS ONE. 2009;4:e6767.
Maeda K, Yamada H, Munetsuna E, Fujii R, Yamazaki M, Ando Y, et al. Association of drinking behaviors with TXNIP DNA methylation levels in leukocytes among the general Japanese population. Am J Drug Alcohol Abus. 2022;48:302–10.
Liu C, Marioni RE, Hedman ÅK, Pfeiffer L, Tsai PC, Reynolds LM, et al. A DNA methylation biomarker of alcohol consumption. Mol Psychiatry. 2018;23:422–33.
Maeda K, Yamada H, Munetsuna E, Fujii R, Yamazaki M, Ando Y, et al. Association of smoking habits with TXNIP DNA methylation levels in leukocytes among general Japanese population. PLoS ONE. 2020;15:e0235486.
Zhang FF, Cardarelli R, Carroll J, Zhang S, Fulda KG, Gonzalez K, et al. Physical activity and global genomic DNA methylation in a cancer-free population. Epigenetics 2011;6:293–9.
Fujii R, Sato S, Tsuboi Y, Cardenas A, Suzuki K. DNA methylation as a mediator of associations between the environment and chronic diseases: A scoping review on application of mediation analysis. Epigenetics. 2022;17:759–85.
Shelnutt KP, Kauwell GP, Gregory JF 3rd, Maneval DR, Quinlivan EP, Theriaque DW, et al. Methylenetetrahydrofolate reductase 677C->T polymorphism affects DNA methylation in response to controlled folate intake in young women. J Nutr Biochem. 2004;15:554–60.
Basten GP, Duthie SJ, Pirie L, Vaughan N, Hill MH, Powers HJ. Sensitivity of markers of DNA stability and DNA repair activity to folate supplementation in healthy volunteers. Br J Cancer. 2006;94:1942–7.
Christensen BC, Kelsey KT, Zheng S, Houseman EA, Marsit CJ, Wrensch MR, et al. Breast cancer DNA methylation profiles are associated with tumor size and alcohol and folate intake. PLoS Genet. 2010;6:e1001043.
Kok DE, Dhonukshe-Rutten RA, Lute C, Heil SG, Uitterlinden AG, van der Velde N, et al. The effects of long-term daily folic acid and vitamin B12 supplementation on genome-wide DNA methylation in elderly subjects. Clin Epigenet. 2015;7:121.
Hibler E, Huang L, Andrade J, Spring B. Impact of a diet and activity health promotion intervention on regional patterns of DNA methylation. Clin Epigenet. 2019;11:133.
Perfilyev A, Dahlman I, Gillberg L, Rosqvist F, Iggman D, Volkov P, et al. Impact of polyunsaturated and saturated fat overfeeding on the DNA-methylation pattern in human adipose tissue: a randomized controlled trial. Am J Clin Nutr. 2017;105:991–1000.
Fujii R, Yamada H, Munetsuna E, Yamazaki M, Mizuno G, Tsuboi Y, et al. Dietary vegetable intake is inversely associated with ATP-binding cassette protein A1 (ABCA1) DNA methylation levels among Japanese women. Nutrition. 2019;65:1–5.
Fujii R, Yamada H, Munetsuna E, Yamazaki M, Ando Y, Mizuno G, et al. Associations between dietary vitamin intake, ABCA1 gene promoter DNA methylation, and lipid profiles in a Japanese population. Am J Clin Nutr. 2019;110:1213–9.
Tsuboi Y, Yamada H, Munetsuna E, Fujii R, Yamazaki M, Ando Y, et al. Intake of vegetables and fruits rich in provitamin A is positively associated with aryl hydrocarbon receptor repressor DNA methylation in a Japanese population. Nutr Res. 2022;107:206–17.
Gibbs JR, van der Brug MP, Hernandez DG, Traynor BJ, Nalls MA, Lai SL, et al. Abundant quantitative trait loci exist for DNA methylation and gene expression in human brain. PLoS Genet. 2010;6:e1000952.
Grundberg E, Meduri E, Sandling JK, Hedman AK, Keildson S, Buil A, et al. Global analysis of DNA methylation variation in adipose tissue from twins reveals links to disease-associated variants in distal regulatory elements. Am J Hum Genet. 2013;93:876–90.
Fujii R, Yamada H, Munetsuna E, Yamazaki M, Mizuno G, Ando Y, et al. Dietary fish and ω-3 polyunsaturated fatty acids are associated with leukocyte ABCA1 DNA methylation levels. Nutrition. 2021;81:110951.
Zhang X, Ritonja JA, Zhou N, Chen BE, Li X. Omega-3 Polyunsaturated Fatty Acids Intake and Blood Pressure: A Dose-Response Meta-Analysis of Randomized Controlled Trials. J Am Heart Assoc. 2022;11:e025071.
Harris WS, Tintle NL, Imamura F, Qian F, Korat AVA, Marklund M, et al. Blood n-3 fatty acid levels and total and cause-specific mortality from 17 prospective studies. Nat Commun. 2021;12:2329.
de Souza RJ, Mente A, Maroleanu A, Cozma AI, Ha V, Kishibe T, et al. Intake of saturated and trans unsaturated fatty acids and risk of all cause mortality, cardiovascular disease, and type 2 diabetes: systematic review and meta-analysis of observational studies. BMJ. 2015;351:h3978.
Mazidi M, Mikhailidis DP, Sattar N, Toth PP, Judd S, Blaha MJ, et al. Association of types of dietary fats and all-cause and cause-specific mortality: A prospective cohort study and meta-analysis of prospective studies with 1,164,029 participants. Clin Nutr. 2020;39:3677–86.
Guay SP, Brisson D, Munger J, Lamarche B, Gaudet D, Bouchard L. ABCA1 gene promoter DNA methylation is associated with HDL particle profile and coronary artery disease in familial hypercholesterolemia. Epigenetics. 2012;7:464–72.
Guay SP, Légaré C, Houde AA, Mathieu P, Bossé Y, Bouchard L. Acetylsalicylic acid, aging and coronary artery disease are associated with ABCA1 DNA methylation in men. Clin Epigenet. 2014;6:14.
Zwarts KY, Clee SM, Zwinderman AH, Engert JC, Singaraja R, Loubser O, et al. ABCA1 regulatory variants influence coronary artery disease independent of effects on plasma lipid levels. Clin Genet. 2002;61:115–25.
Ma Y, Follis JL, Smith CE, Tanaka T, Manichaikul AW, Chu AY, et al. Interaction of methylation-related genetic variants with circulating fatty acids on plasma lipids: a meta-analysis of 7 studies and methylation analysis of 3 studies in the Cohorts for Heart and Aging Research in Genomic Epidemiology consortium. Am J Clin Nutr. 2016;103:567–78.
Tokudome S, Goto C, Imaeda N, Tokudome Y, Ikeda M, Maki S. Development of a data-based short food frequency questionnaire for assessing nutrient intake by middle-aged Japanese. Asian Pac J Cancer Prev. 2004;5:40–3.
Imaeda N, Goto C, Sasakabe T, Mikami H, Oze I, Hosono A, et al. Reproducibility and validity of food group intake in a short food frequency questionnaire for the middle-aged Japanese population. Environ Health Prev Med. 2021;26:28.
Tokudome Y, Goto C, Imaeda N, Hasegawa T, Kato R, Hirose K, et al. Relative validity of a short food frequency questionnaire for assessing nutrient intake versus three-day weighed diet records in middle-aged Japanese. J Epidemiol. 2005;15:135–45.
Goto C, Tokudome Y, Imaeda N, Takekuma K, Kuriki K, Igarashi F, et al. Validation study of fatty acid consumption assessed with a short food frequency questionnaire against plasma concentration in middle-aged Japanese people. Scand J Food Nutr. 2006;50:77–82.
Willett WC, Howe GR, Kushi LH. Adjustment for total energy intake in epidemiologic studies. Am J Clin Nutr. 1997;65:1220S–8S.
Hardy GH. Mendelian proportions in a mixed population. Science. 1908;28:49–50.
Weinberg W. Über den Nachweis der Vererbung beim Menschen Jahresh (About proof of heredity in humans). Ver f Vater Nat Wüttemberg. 1908;64:368–82.
Qiu W, Wan E, Morrow J, Cho MH, Crapo JD, Silverman EK, et al. The impact of genetic variation and cigarette smoke on DNA methylation in current and former smokers from the COPDGene study. Epigenetics. 2015;10:1064–73.
Breitling LP. Current genetics and epigenetics of smoking/tobacco-related cardiovascular disease. Arterioscler Thromb Vasc Biol. 2013;33:1468–72.
Gao X, Thomsen H, Zhang Y, Breitling LP, Brenner H. The impact of methylation quantitative trait loci (mQTLs) on active smoking-related DNA methylation changes. Clin Epigenet. 2017;9:87.
Zeng Y, Amador C, Gao C, Walker RM, Morris SW, Campbell A, et al. Lifestyle and Genetic Factors Modify Parent-of-Origin Effects on the Human Methylome. EBioMedicine. 2021;74:103730.
Wang T, Heianza Y, Sun D, Zheng Y, Huang T, Ma W, et al. Improving fruit and vegetable intake attenuates the genetic association with long-term weight gain. Am J Clin Nutr. 2019;110:759–68.
Chen Y, Liu H, Wang L, Zhou T, Liang Z, Li W, et al. Lifestyle intervention modifies the effect of the MC4R genotype on changes in insulin resistance among women with prior gestational diabetes: Tianjin Gestational Diabetes Mellitus Prevention Program. Am J Clin Nutr. 2019;110:750–8.
Hüls A, Wright MN, Bogl LH, Kaprio J, Lissner L, Molnár D, et al. Polygenic risk for obesity and its interaction with lifestyle and sociodemographic factors in European children and adolescents. Int J Obes (Lond). 2021;45:1321–30.
Brahe LK, Ängquist L, Larsen LH, Vimaleswaran KS, Hager J, Viguerie N, et al. Influence of SNPs in nutrient-sensitive candidate genes and gene-diet interactions on blood lipids: the DiOGenes study. Br J Nutr. 2013;110:790–6.
InterAct Consortium. Investigation of gene-diet interactions in the incretin system and risk of type 2 diabetes: the EPIC-InterAct study. Diabetologia. 2016;59:2613–21.
Li SX, Imamura F, Ye Z, Schulze MB, Zheng J, Ardanaz E, et al. Interaction between genes and macronutrient intake on the risk of developing type 2 diabetes: systematic review and findings from European Prospective Investigation into Cancer (EPIC)-InterAct. Am J Clin Nutr. 2017;106:263–75.
Livingstone KM, Abbott G, Bowe SJ, Ward J, Milte C, McNaughton SA. Diet quality indices, genetic risk and risk of cardiovascular disease and mortality: a longitudinal analysis of 77 004 UK Biobank participants. BMJ Open. 2021;11:e045362.
Rutten-Jacobs LC, Larsson SC, Malik R, Rannikmäe K, MEGASTROKE consortium; International Stroke Genetics Consortium, Sudlow CL, et al. Genetic risk, incident stroke, and the benefits of adhering to a healthy lifestyle: cohort study of 306 473 UK Biobank participants. BMJ. 2018;363:k4168.
Aslibekyan S, Wiener HW, Havel PJ, Stanhope KL, O'Brien DM, Hopkins SE, et al. DNA methylation patterns are associated with n-3 fatty acid intake in Yup'ik people. J Nutr. 2014;144:425–30.
Tremblay BL, Guénard F, Rudkowska I, Lemieux S, Couture P, Vohl MC. Epigenetic changes in blood leukocytes following an omega-3 fatty acid supplementation. Clin Epigenetics. 2017;9:43.
Acknowledgements
We are grateful to residents for participation in our study and to staff of the Health Examination Program for Residents for their support.
Funding
This study was supported by the JSPS KAKENHI Grants-in-Aid for Scientific Research (Nos. 26293144 and 17K09139), Grants-in-Aid for Scientific Research on Innovative Areas [CoBiA] (Nos. 16H06277 22H04923), and the Suzuken Memorial Foundation (No. 18-031).
Author information
Authors and Affiliations
Contributions
KS and KW designed the health checkup; RF, YA, HY, YT, MY, GM, KO, HI, and KS contributed for data acquisition; YA, HY, EM, MY, and GM conducted genotyping and measured methylation levels; NI and CG were responsible for nutritional survey; RF and YA analyzed data and wrote the paper; KM and MW reviewed the paper critically for important intellectual content; and KS and RF had primary responsibility for final content. All authors reviewed and approved the final paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
This study was conducted in accordance with the Declaration of Helsinki. The study protocol was approved by the Ethics Committee of Fujita Health University (No. HG19-069).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Fujii, R., Ando, Y., Yamada, H. et al. Integration of methylation quantitative trait loci (mQTL) on dietary intake on DNA methylation levels: an example of n-3 PUFA and ABCA1 gene. Eur J Clin Nutr 77, 881–887 (2023). https://doi.org/10.1038/s41430-023-01315-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41430-023-01315-6