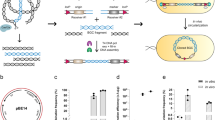
Abstract
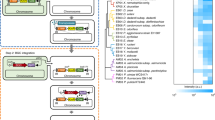
The biosynthetic gene clusters (BGCs) for the macrocyclic lactone-based polyketide compounds are extremely large-sized because the polyketide synthases that generate the polyketide chains of the basic backbone are of very high molecular weight. In developing a heterologous expression system for the large BGCs amenable to the production of such natural products, we selected concanamycin as an appropriate target. We obtained a bacterial artificial chromosome (BAC) clone with a 211-kb insert harboring the entire BGC responsible for the biosynthesis of concanamycin. Heterologous expression of this clone in a host strain, Streptomyces avermitilis SUKA32, permitted the production of concanamycin, as well as that of two additional aromatic polyketides. Structural elucidation identified these additional products as ent-gephyromycin and a novel compound that was designated JBIR-157. We describe herein sequencing and expression studies performed on these BGCs, demonstrating the utility of large BAC clones for the heterologous expression of cryptic or near-silent loci.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Pye CR, Bertin MJ, Lokey RS, Gerwick WH, Linington RG. Retrospective analysis of natural products provides insights for future discovery trends. Proc Natl Acad Sci USA. 2017;114:5601–6.
Palazzolo AME, Simons CLW, Burke MD. The natural productome. Proc Natl Acad Sci USA. 2017;114:5564–6.
Singh V, Haque S, Niwas R, Srivastava A, Pasupuleti M, Tripathi CK. Strategies for fermentation medium optimization: an in-depth review. Front Microbiol. 2016;7:2087.
Hoshino S, Onaka H, Abe I. Activation of silent biosynthetic pathways and discovery of novel secondary metabolites in actinomycetes by co-culture with mycolic acid-containing bacteria. J Ind Microbiol Biotechnol. 2019;46:363–74.
Adrio JL, Demain AL. Genetic improvement of processes yielding microbial products. FEMS Microbiol Rev. 2006;30:187–214.
Cimermancic P, Medema MH, Claesen J, Kurita K, Wieland Brown LC, Mavrommatis K, et al. Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cell. 2014;158:412–21.
Doroghazi JR, Albright JC, Goering AW, Ju KS, Haines RR, Tchalukov KA, et al. A roadmap for natural product discovery based on large-scale genomics and metabolomics. Nat Chem Biol. 2014;10:963–8.
Komatsu M, Komatsu K, Koiwai H, Yamada Y, Kozone I, Izumikawa M, et al. Engineered Streptomyces avermitilis host for heterologous expression of biosynthetic gene cluster for secondary metabolites. ACS Synth Biol. 2013;2:384–96.
Hashimoto T, Hashimoto J, Kozone I, Amagai K, Kawahara T, Takahashi S, et al. Biosynthesis of quinolidomicin, the largest known macrolide of terrestrial origin: identification and heterologous expression of a biosynthetic gene cluster over 200 kb. Org Lett. 2018;20:7996–9.
Zhang L, Hashimoto T, Qin B, Hashimoto J, Kozone I, Kawahara T, et al. Characterization of giant modular PKSs provides insight into genetic mechanism for structural diversification of aminopolyol polyketides. Angew Chem Int Ed Engl. 2017;56:1740–5.
Hashimoto T, Kozone I, Hashimoto J, Ueoka R, Kagaya N, Fujie M, et al. Novel macrolactam compound produced by the heterologous expression of a large cryptic biosynthetic gene cluster of Streptomyces rochei IFO12908. J Antibiot. 2020;73:171–4.
Hashimoto T, Kozone I, Hashimoto J, Suenaga H, Fujie M, Satoh N, et al. Identification, cloning and heterologous expression of biosynthetic gene cluster for desertomycin. J Antibiot. 2020;73:650–4.
Kinashi H, Someno K, Sakaguchi K, Higashijima T, Miyazawa T. Structure of concanamycin a. Tetrahedron Lett. 1981;22:3861–4.
Kinashi H, Someno K, Sakaguchi K. Isolation and characterization of concanamycins A, B and C. J Antibiot. 1984;37:1333–43.
Yamamoto H, Nakazawa K, Horii S, Miyake A. Studies on Agricultural Antibiotic. J Agric Chem Soc Jpn. 1960;34:268–72.
Drose S, Bindseil KU, Bowman EJ, Siebers A, Zeeck A, Altendorf K. Inhibitory effect of modified bafilomycins and concanamycins on P- and V-type adenosinetriphosphatases. Biochemistry. 1993;32:3902–6.
Haydock SF, Appleyard AN, Mironenko T, Lester J, Scott N, Leadlay PF. Organization of the biosynthetic gene cluster for the macrolide concanamycin A in Streptomyces neyagawaensis ATCC 27449. Microbiology. 2005;151:3161–9.
Demachi A, Ohte S, Uchida R, Shin-Ya K, Ohshiro T, Tomoda H, et al. Discovery of prescopranone, a key intermediate in scopranone biosynthesis. J Antibiot. 2022;75:305–11.
Cane DE, He X, Kobayashi S, Ōmura S, Ikeda H. Geosmin biosynthesis in Streptomyces avermitilis. Molecular cloning, expression, and mechanistic study of the germacradienol/geosmin synthase. J Antibiot. 2006;59:471–9.
Komatsu M, Uchiyama T, Ōmura S, Cane DE, Ikeda H. Genome-minimized Streptomyces host for the heterologous expression of secondary metabolism. Proc Natl Acad Sci USA. 2010;107:2646–51.
Kim JH, Komatsu M, Shin-ya K, Ōmura S, Ikeda H. Distribution and functional analysis of the phosphopantetheinyl transferase superfamily in Actinomycetales microorganisms. Proc Natl Acad Sci USA. 2018;115:6828–33.
Chu G, Vollrath D, Davis RW. Separation of large DNA molecules by contour-clamped homogeneous electric fields. Science. 1986;234:1582–5.
Sievers F, Higgins DG. Clustal Omega for making accurate alignments of many protein sequences. Protein Sci. 2017;27:135–45.
Bringmann G, Lang G, Maksimenka K, Hamm A, Gulder TA, Dieter A, et al. Gephyromycin, the first bridged angucyclinone, from Streptomyces griseus strain NTK 14. Phytochemistry. 2005;66:1366–73.
Chang Y, Xing L, Sun C, Liang S, Liu T, Zhang X, et al. Monacycliones G–K and ent-gephyromycin A, angucycline derivatives from the marine-derived Streptomyces sp. HDN15129. J Nat Prod. 2020;83:2749–55.
Keatinge-Clay A. Crystal structure of the erythromycin polyketide synthase dehydratase. J Mol Biol. 2008;384:941–53.
Zhang W, Fortman JL, Carlson JC, Yan J, Liu Y, Bai F, et al. Characterization of the bafilomycin biosynthetic gene cluster from Streptomyces lohii. ChemBioChem. 2013;14:301–6.
Zhu W-Z, Wang S-H, Gao H-M, Ge Y-M, Dai J, Zhang X-L, et al. Characterization of bioactivities and biosynthesis of angucycline/angucyclinone derivatives derived from Gephyromycinifex aptenodytis gen. nov., sp. nov. Mar Drugs. 2021;20:34.
Davis NK, Chater KF. Spore colour in Streptomyces coelicolor A3(2) involves the developmentally regulated synthesis of a compound biosynthetically related to polyketide antibiotics. Mol Microbiol. 1990;4:1679–91.
Kautsar SA, Blin K, Shaw S, Navarro-Munoz JC, Terlouw BR, van der Hooft JJJ, et al. MIBiG 2.0: a repository for biosynthetic gene clusters of known function. Nucleic Acids Res. 2020;48:D454–D8.
McDaniel R, Ebert-Khosla S, Hopwood DA, Khosla C. Engineered biosynthesis of novel polyketides. Science. 1993;262:1546–50.
Kulowski K, Wendt-Pienkowski E, Han L, Yang K, Vining LC, Hutchinson CR. Functional characterization of the jadI gene as a cyclase forming angucyclinones. J Am Chem Soc. 1999;121:1786–94.
Westrich L, Domann S, Faust B, Bedford D, Hopwood DA, Bechthold A. Cloning and characterization of a gene cluster from Streptomyces cyanogenus S136 probably involved in landomycin biosynthesis. FEMS Microbiol Lett. 1999;170:381–7.
Decker H, Haag S. Cloning and characterization of a polyketide synthase gene from Streptomyces fradiae Tu2717, which carries the genes for biosynthesis of the angucycline antibiotic urdamycin A and a gene probably involved in its oxygenation. J Bacteriol. 1995;177:6126–36.
Han L, Yang K, Ramalingam E, Mosher RH, Vining LC. Cloning and characterization of polyketide synthase genes for jadomycin B biosynthesis in Streptomyces venezuelae ISP5230. Microbiology. 1994;140:3379–89.
Patrikainen P, Kallio P, Fan K, Klika KD, Shaaban KA, Mantsala P, et al. Tailoring enzymes involved in the biosynthesis of angucyclines contain latent context-dependent catalytic activities. Chem Biol. 2012;19:647–55.
Kallio P, Patrikainen P, Suomela JP, Mantsala P, Metsa-Ketela M, Niemi J. Flavoprotein hydroxylase PgaE catalyzes two consecutive oxygen-dependent tailoring reactions in angucycline biosynthesis. Biochemistry. 2011;50:5535–43.
Künzel E, Faust B, Oelkers C, Weissbach U, Bearden DW, Weitnauer G, et al. Inactivation of the urdGT2 gene, which encodes a glycosyltransferase responsible for the C-glycosyltransfer of activated d-olivose, leads to formation of the novel urdamycins I, J, and K. J Am Chem Soc. 1999;121:11058–62.
Palmu K, Ishida K, Mantsala P, Hertweck C, Metsa-Ketela M. Artificial reconstruction of two cryptic angucycline antibiotic biosynthetic pathways. ChemBioChem. 2007;8:1577–84.
Sasaki T, Gomi S, Sezaki M, Takeuchi Y, Kodama Y, Kawamura K. New antibiotics SF2315A and B produced by an Excellospora sp. II. The structural elucidation. J Antibiot. 1988;41:843–8.
Ishikawa K, Hashimoto M, Komatsu K, Taguchi T, Okamoto S, Ichinose K. Characterization of stereospecific enoyl reductase ActVI-ORF2 for pyran ring formation in the actinorhodin biosynthesis of Streptomyces coelicolor A3(2). Bioorg Med Chem Lett. 2022;66:128727.
Sasaki E, Ogasawara Y, Liu HW. A biosynthetic pathway for BE-7585A, a 2-thiosugar-containing angucycline-type natural product. J Am Chem Soc. 2010;132:7405–17.
Schafer M, Le TB, Hearnshaw SJ, Maxwell A, Challis GL, Wilkinson B, et al. SimC7 is a novel NAD(P)H-dependent ketoreductase essential for the antibiotic activity of the DNA gyrase inhibitor simocyclinone. J Mol Biol. 2015;427:2192–204.
Sciara G, Kendrew SG, Miele AE, Marsh NG, Federici L, Malatesta F, et al. The structure of ActVA-Orf6, a novel type of monooxygenase involved in actinorhodin biosynthesis. EMBO J. 2003;22:205–15.
Takao R, Sakai K, Koshino H, Osada H, Takahashi S. Identification of the kinanthraquinone biosynthetic gene cluster by expression of an atypical response regulator. Biosci Biotechnol Biochem. 2021;85:714–21.
Acknowledgements
MI deceased on 23 December 2015. This work was supported by AMED under Grant JP19ae0101045 to KS and JSPS KAKENHI Grant Number JP23H04569 to KK.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kudo, K., Nishimura, T., Izumikawa, M. et al. Capability of a large bacterial artificial chromosome clone harboring multiple biosynthetic gene clusters for the production of diverse compounds. J Antibiot 77, 288–298 (2024). https://doi.org/10.1038/s41429-024-00711-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41429-024-00711-9