Abstract
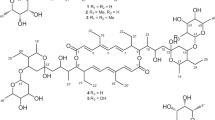
Ten new proansamycin B congeners (1–10) together with one known (11) were isolated and characterized on the basis of 1D and 2D NMR spectroscopic and HRESIMS data from the Amycolatopsis mediterranei S699 ΔPM::rifR+rif-orf19 mutant. Compounds 8 and 9 featured with six-membered ring and five-membered ring hemiketal, respectively. Compounds 1, 2, and 9 displayed antibacterial activity against MRSA (methicillin-resistant Staphylococcus aureus), with the MIC (minimal inhibitory concentration) values of 64, 8, and 128 µg/mL, respectively. Compound 1 showed significant cytotoxicity against MDA-MB-231, HepG2 and Panc-1 cell lines with IC50 (half maximal inhibitory concentration) values of 2.3 ± 0.2, 2.5 ± 0.3 and 3.8 ± 0.5 μM, respectively.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Floss HG, Yu TW. Rifamycin-mode of action, resistance, and biosynthesis. Chem Rev. 2005;105:621–32.
Kupchan SM, Komoda Y, Court WA, Thomas GJ, Smith RM, Karim A, et al. Maytansine, a novel antileukemic ansa macrolide from Maytenus ovatus. J Am Chem Soc. 1972;94:1354–6.
Sasaki K, Rinehart KL Jr, Slomp G, Grostic MF, Olson EC. Geldanamycin. I. structure assignment. J Am Chem Soc. 1970;92:7591–3.
Kang Q, Shen Y, Bai L. Biosynthesis of 3,5-AHBA-derived natural products. Nat Prod Rep. 2012;29:243–63.
Campbell EA, Korzheva N, Mustaev A, Murakami K, Nair S, Goldfarb A, et al. Structural mechanism for rifampicin inhibition of bacterial rna polymerase. Cell. 2001;104:901–12.
Artsimovitch I, Vassylyeva MN, Svetlov D, Svetlov V, Perederina A, Igarashi N, et al. Allosteric modulation of the RNA polymerase catalytic reaction is an essential component of transcription control by rifamycins. Cell. 2005;122:351–63.
Ramaswamy S, Musser JM. Molecular genetic basis of antimicrobial agent resistance in Mycobacterium tuberculosis: 1998 update. Tuber Lung Dis. 1998;79:3–29.
Siu GK, Zhang Y, Lau TC, Lau RW, Ho PL, Yew WW, et al. Mutations outside the rifampicin resistance-determining region associated with rifampicin resistance in Mycobacterium tuberculosis. J Antimicrob Chemothe. 2011;66:730–3.
Ye F, Zhao X, Shi Y, Hu Y, Ding Y, Lu C, et al. Deciphering the timing of naphthalenic ring formation in the Biosynthesis of 8-Deoxyrifamycins. Org Lett. 2023;25:6474–78.
Han TY, Zhang K, Tang GL, Zhou Q. Characterizing Post‐PKS Modifications of 16‐Demethyl‐rifamycin revealed two dehydrogenases diverting the aromatization mode of naphthalenic ring in ansamycin biosynthesis. Chin J Chem. 2022;10:9–15.
Ye F, Shi Y, Zhao S, Li Z, Wang H, Lu C, et al. 8-Deoxy-Rifamycin Derivatives from Amycolatopsis mediterranei S699 Delta rifT Strain. Biomolecules. 2020;10:1265.
Shi YR, Ye F, Song YL, Zhang XC, Lu CH, Shen YM. Rifamycin W Analogues from Amycolatopsis mediterranei S699 Delta rif-orf5 Strain. Biomolecules. 2021;11:920.
Xu J, Wan E, Kim CJ, Floss HG, Mahmud T. Identification of tailoring genes involved in the modification of the polyketide backbone of rifamycin B by Amycolatopsis mediterranei S699. Microbioloogy. 2005;151:2515–28.
Qi F, Lei C, Li F, Zhang X, Wang J, Zhang W, et al. Deciphering the late steps of rifamycin biosynthesis. Nat Commun. 2018;9:2342.
Stratmann A, Schupp T, Toupet C, Schilling W, Oberer L, Traber R. New insights into rifamycin B biosynthesis: isolation of proansamycin B and 34a-deoxy-rifamycin W as early macrocyclic intermediates indicating two separated biosynthetic pathways. J Antibiot. 2002;55:396–406.
Hutchinson CR, Floss HG. Biosynthesis of the ansamycin antibiotic rifamycin: deductions from the molecular analysis of the rif biosynthetic gene cluster of Amycolatopsis mediterranei S699. Chem Biol. 1998;5:69–79.
Ghisalba O, Traxler P, Fuhrer H, Richter WJ. Early intermediates in the biosynthesis of ansamycins. II. Isolation and identification of proansamycin B-M1 and protorifamycin I-M1. J Antibiot. 1979;32:1267–72.
Zhang J, Li S, Wu X, Guo Z, Lu C, Shen Y. Nam7 hydroxylase is responsible for the formation of the naphthalenic ring in the biosynthesis of neoansamycins. Org Lett. 2017;19:2442–45.
Zhou Q, Luo G-C, Zhang H, Tang G-L. 34a-Hydroxylation in Rifamycin biosynthesis catalyzed by cytochrome P450 encoded by rif-orf13. Chin J Org Chem. 2019;39:58–63.
Shi Y, Zhang J, Tian X, Wu X, Li T, Lu C, et al. Isolation of 11,12-seco-Rifamycin W Derivatives Reveals a Cleavage Pattern of the Rifamycin Ansa Chain. Org Lett. 2019;21:900–03.
Li S, Lu C, Ou J, Deng J, Shen Y. Overexpression of hgc1 increases the production and diversity of hygrocins in Streptomyces sp. LZ35. RSC Adv. 2015;5:83843–46.
Zhao GS, Li SR, Guo ZX, Sun MW, Lu CH. Overexpression of div8 increases the production and diversity of divergolides in Streptomyces sp W112. Rsc Adv. 2015;5:98209–14.
Wang J, Li W, Wang H, Lu C. Pentaketide Ansamycin Microansamycins A-I from Micromonospora sp. Org Lett. 2018;20:1058–61.
Xiao YS, Zhang B, Zhang M, Guo ZK, Deng XZ, Shi J, et al. Rifamorpholines A–E, potential antibiotics from locust-associated actinobacteria Amycolatopsis sp. Hca4. Org Biomol Chem. 2017;15:3909–16.
Funding
This research was supported by the National Key Research and Development Program (2019YFA0905402), the National Natural Science Foundation of China (81673317, 81602979, 81530091), the Program for Changjiang Scholars and Innovative Research Team in University (IRT_17R68).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ma, X., Ye, F., Zhang, X. et al. Proansamycin B derivatives from the post-PKS modification gene deletion mutant of Amycolatopsis mediterranei S699. J Antibiot 77, 278–287 (2024). https://doi.org/10.1038/s41429-024-00708-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41429-024-00708-4