Abstract
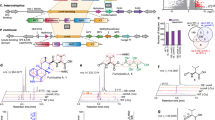
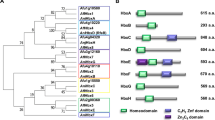
Copper is a transition metal element with significant effects on the morphological development and secondary metabolism of actinobacteria. In some microorganisms, copper-binding natural products are employed to modulate copper homeostasis, although their significance in actinobacteria remains largely unknown. Here, we identified the biosynthetic genes of the diisocyanide natural product hazimycin in Kitasatospora purpeofusca HV058, through gene knock-out and heterologous expression. Biochemical analyses revealed that hazimycin A specifically binds to copper, which diminishes its antimicrobial activity. The presence of a set of putative importer/exporter genes surrounding the biosynthetic genes suggested that hazimycin is a chalkophore that modulates the intracellular copper level. A bioinformatic survey of homologous gene cassettes, as well as the identification of two previously unknown hazimycin-producing Streptomyces strains, indicated that the isocyanide-based mechanism of copper homeostasis is prevalent in actinobacteria.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Genome sequence of K. purpeofusca HV058 was deposited in DDBJ under following accession ID: AP028455 (chromosome) and AP028456 (plasmid).
References
Kenney GE, Rosenzweig AC. Chalkophores. Annu Rev Biochem. 2018;87:645–76.
Johnstone TC, Nolan EM. Beyond iron: non-classical biological functions of bacterial siderophores. Dalton Trans. 2015;44:6320–39.
Kim HJ, Graham DW, DiSpirito AA, Alterman MA, Galeva N, Larive CK, et al. Methanobactin, a copper-acquisition compound from methane-oxidizing bacteria. Science. 2004;305:1612–5.
Kenney GE, Dassama LMK, Pandelia ME, Gizzi AS, Martinie RJ, Gao P, et al. The biosynthesis of methanobactin. Science. 2018;359:1411–16.
Anttila J, Heinonen P, Nenonen T, Pino A, Iwaï H, Kauppi E, et al. Is coproporphyrin III a copper-acquisition compound in Paracoccus denitrificans? Biochim Biophys Acta. 2011;1807:311–8.
Chaturvedi KS, Hung CS, Crowley JR, Stapleton AE, Henderson JP. The siderophore yersiniabactin binds copper to protect pathogens during infection. Nat Chem Biol. 2012;8:731–6.
Koh EI, Robinson AE, Bandara N, Rogers BE, Henderson JP. Copper import in Escherichia coli by the yersiniabactin metallophore system. Nat Chem Biol. 2017;13:1016–21.
Patteson JB, Putz AT, Tao L, Simke WC, Bryant LH 3rd, et al. Biosynthesis of fluopsin C, a copper-containing antibiotic from Pseudomonas aeruginosa. Science. 2021;374:1005–9.
Rothe W. The new antibiotic xanthocillin. Dtsch Med Wochenschr. 1954;79:1080–1.
Edenborough MS, Herbert RB. Naturally occurring isocyanides. Nat Prod Rep. 1988;5:229–45.
Garson MJ, Simpson JS. Marine isocyanides and related natural products-structure, biosynthesis and ecology. Nat Prod Rep. 2004;21:164–79.
Emsermann J, Kauhl U, Opatz T. Marine isonitriles and their related compounds. Mar Drugs. 2016;14:16.
Hohlman RM, Sherman DH. Recent advances in hapalindole-type cyanobacterial alkaloids: biosynthesis, synthesis, and biological activity. Nat Prod Rep. 2021;38:1567–88.
Massarotti A, Brunelli F, Aprile S, Giustiniano M, Tron GC. Medicinal chemistry of isocyanides. Chem Rev. 2021;121:10742–88.
Lim FY, Won TH, Raffa N, Baccile JA, Wisecaver J, Rokas A, et al. Fungal isocyanide synthases and xanthocillin biosynthesis in aspergillus fumigatus. mBio. 2018;9:e00785–18.
Raffa N, Won TH, Sukowaty A, Candor K, Cui C, Halder S, et al. Dual-purpose isocyanides produced by Aspergillus fumigatus contribute to cellular copper sufficiency and exhibit antimicrobial activity. Proc Natl Acad Sci USA. 2021;118:e2015224118.
Hübner I, Shapiro JA, Hoßmann J, Drechsel J, Hacker SM, Rather PN, et al. Broad spectrum antibiotic xanthocillin X effectively kills acinetobacter baumanniivia dysregulation of heme biosynthesis. ACS Cent Sci. 2021;7:488–98.
Wang L, Zhu M, Zhang Q, Zhang X, Yang P, Liu Z, et al. Diisonitrile natural product SF2768 functions as a chalkophore that mediates copper acquisition in Streptomyces thioluteus. ACS Chem Biol. 2017;12:3067–75.
Harris NC, Sato M, Herman NA, Twigg F, Cai W, Liu J, et al. Biosynthesis of isonitrile lipopeptides by conserved nonribosomal peptide synthetase gene clusters in Actinobacteria. Proc Natl Acad Sci USA. 2017;114:7025–30.
Mehdiratta K, Singh S, Sharma S, Bhosale RS, Choudhury R, Masal DP, et al. Kupyaphores are zinc homeostatic metallophores required for colonization of Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2022;119:e2110293119.
Del Rio Flores A, Narayanamoorthy M, Cai W, Zhai R, Yang S, Shen Y, et al. Biosynthesis of isonitrile lipopeptide metallophores from pathogenic mycobacteria. Biochemistry. 2023;62:824–34.
Ueda K, Tomaru Y, Endoh K, Beppu T. Stimulatory effect of copper on antibiotic production and morphological differentiation in Streptomyces tanashiensis. J Antibiot. 1997;50:693–5.
Keijser BJ, van Wezel GP, Canters GW, Kieser T, Vijgenboom E. The ram-dependence of Streptomyces lividans differentiation is bypassed by copper. J Mol Microbiol Biotechnol. 2000;2:565–74.
Worrall JA, Vijgenboom E. Copper mining in Streptomyces: enzymes, natural products and development. Nat Prod Rep. 2010;27:742–56.
Locatelli FM, Goo KS, Ulanova D. Effects of trace metal ions on secondary metabolism and the morphological development of Streptomycetes. Metallomics. 2016;8:469–80.
González-Quiñónez N, Corte-Rodríguez M, Álvarez-Fernández-García R, Rioseras B, López-García MT, Fernández-García G, et al. Cytosolic copper is a major modulator of germination, development and secondary metabolism in Streptomyces coelicolor. Sci Rep. 2019;9:4214. 12
Kim Wright JJ, Cooper AB, McPhail AT, Merrill Y, Nagabhushan TL, Puar MS. X-Ray crystal structure determination and synthesis of the new isonitrile-containing antibiotics, hazimycin factors 5 and 6. J Chem Soc Chem Commun. 1982;1188–90.
Marquez JA, Horan AC, Kalyanpur M, Lee BK, Loebenberg D, Miller GH, et al. The hazimicins, a new class of antibiotics. Taxonomy, fermentation, isolation, characterization and biological properties. J Antibiot. 1983;36:1101–8.
Koyama N, Sato H, Tomoda H. Discovery of new hazimycin congeners from Kitasatospora sp. P07101. Acta Pharm Sin B. 2015;5:564–8.
Briza P, Eckerstorfer M, Breitenbach M. The sporulation-specific enzymes encoded by the DIT1 and DIT2 genes catalyze a two-step reaction leading to a soluble LL-dityrosine-containing precursor of the yeast spore wall. Proc Natl Acad Sci USA. 1994;91:4524–8.
Brady SF, Clardy J. Cloning and heterologous expression of isocyanide biosynthetic genes from environmental DNA. Angew Chem Int Ed Engl. 2005;44:7063–5.
Drake EJ, Gulick AM. Three-dimensional structures of Pseudomonas aeruginosa PvcA and PvcB, two proteins involved in the synthesis of 2-isocyano-6,7-dihydroxycoumarin. J Mol Biol. 2008;384:193–205.
Crawford JM, Kontnik R, Clardy J. Regulating alternative lifestyles in entomopathogenic bacteria. Curr Biol. 2010;20:69–74.
Huang YB, Cai W, Del Rio Flores A, Twigg FF, Zhang W. Facile discovery and quantification of isonitrile natural products via tetrazine-based click reactions. Anal Chem. 2020;92:599–602.
Funabashi M, Yang Z, Nonaka K, Hosobuchi M, Fujita Y, Shibata T, et al. An ATP-independent strategy for amide bond formation in antibiotic biosynthesis. Nat Chem Biol. 2010;6:581–6.
Goda M, Hashimoto Y, Shimizu S, Kobayashi M. Discovery of a novel enzyme, isonitrile hydratase, involved in nitrogen-carbon triple bond cleavage. J Biol Chem. 2001;276:23480–5.
Sato H, Hashimoto Y, Fukatsu H, Kobayashi M. Novel isonitrile hydratase involved in isonitrile metabolism. J Biol Chem. 2010;285:34793–802.
Hu W, Zheng H. Cryo-EM reveals unique structural features of the FhuCDB Escherichia coli ferrichrome importer. Commun Biol. 2021;4:1383.
Korkhov VM, Mireku SA, Locher KP. Structure of AMP-PNP-bound vitamin B12 transporter BtuCD-F. Nature. 2012;490:367–72.
Choi DW, Zea CJ, Do YS, Semrau JD, Antholine WE, Hargrove MS, et al. Spectral, kinetic, and thermodynamic properties of Cu(I) and Cu(II) binding by methanobactin from Methylosinus trichosporium OB3b. Biochemistry. 2006;45:1442–53.
Zhu M, Wang L, Zhang W, Liu Z, Ali M, Imtiaz M, He J. Diisonitrile-mediated reactive oxygen species accumulation leads to bacterial growth inhibition. J Nat Prod. 2020;83:1634–40.
Srinivasan S, Avadhani NG. Cytochrome c oxidase dysfunction in oxidative stress. Free Radic Biol Med. 2012;53:1252–63.
Navarro-Muñoz JC, Selem-Mojica N, Mullowney MW, Kautsar SA, Tryon JH, Parkinson EI, et al. A computational framework to explore large-scale biosynthetic diversity. Nat Chem Biol. 2020;16:60–68.
Brady SF, Clardy J. Systematic investigation of the Escherichia coli metabolome for the biosynthetic origin of an isocyanide carbon atom. Angew Chem Int Ed Engl. 2005;44:7045–8.
Chang WC, Sanyal D, Huang JL, Ittiamornkul K, Zhu Q, Liu X. In vitro stepwise reconstitution of amino acid derived vinyl isocyanide biosynthesis: detection of an elusive intermediate. Org Lett. 2017;19:1208–11.
Harris NC, Born DA, Cai W, Huang Y, Martin J, Khalaf R, et al. Isonitrile formation by a non-heme iron(ii)-dependent oxidase/decarboxylase. Angew Chem Int Ed Engl. 2018;57:9707–10.
Dose B, Niehs SP, Scherlach K, Shahda S, Flórez LV, Kaltenpoth M, et al. Biosynthesis of Sinapigladioside, an Antifungal Isothiocyanate from Burkholderia Symbionts. Chembiochem. 2021;22:1920–24.
Won TH, Bok JW, Nadig N, Venkatesh N, Nickles G, Greco C, et al. Copper starvation induces antimicrobial isocyanide integrated into two distinct biosynthetic pathways in fungi. Nat Commun. 2022;13:4828.
Chen TY, Zheng Z, Zhang X, Chen J, Cha L, Tang Y, et al. Deciphering the reaction pathway of mononuclear iron enzyme-catalyzed N≡C triple bond formation in isocyanide lipopeptide and polyketide biosynthesis. ACS Catal. 2022;12:2270–79.
Hayakawa M, Nonomura H. Humic acid-vitamin agar, a new medium for the selective isolation of soil actinomycetes. J Ferment. 1987;65:501–9.
Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical Streptomyces genetics. Norwich: John Innes Foundation; 2000.
Kolmogorov M, Yuan J, Lin Y, Pevzner PA. Assembly of long, error-prone reads using repeat graphs. Nat Biotechnol. 2019;37:540–6.
Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE. 2014;9:e112963.
Vaser R, Sović I, Nagarajan N, Šikić M. Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res. 2017;27:737–46.
Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–55.
Tanizawa Y, Fujisawa T, Kaminuma E, Nakamura Y, Arita M. DFAST and DAGA: web-based integrated genome annotation tools and resources. Biosci Microbiota Food Health. 2016;35:173–84.
Chaumeil PA, Mussig AJ, Hugenholtz P, Parks DH. GTDB-Tk v2: memory friendly classification with the genome taxonomy database. Bioinformatics. 2022;38:5315–16.
Feeney MA, Newitt JT, Addington E, Algora-Gallardo L, Allan C, Balis L, et al. ActinoBase: tools and protocols for researchers working on Streptomyces and other filamentous actinobacteria. Micro Genom. 2022;8:mgen000824.
Kim JH, Komatsu M, Shin-Ya K, Omura S, Ikeda H. Distribution and functional analysis of the phosphopantetheinyl transferase superfamily in Actinomycetales microorganisms. Proc Natl Acad Sci USA. 2018;115:6828–33.
Komatsu M, Uchiyama T, Omura S, Cane DE, Ikeda H. Genome-minimized Streptomyces host for the heterologous expression of secondary metabolism. Proc Natl Acad Sci USA. 2010;107:2646–51.
Matsuda K, Kobayashi M, Kuranaga T, Takada K, Ikeda H, Matsunaga S, et al. SurE is a trans-acting thioesterase cyclizing two distinct non-ribosomal peptides. Org Biomol Chem. 2019;17:1058–61.
Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, et al. BLAST+: architecture and applications. BMC Bioinforma. 2009;10:421.
Eddy SR. Accelerated profile HMM searches. PLoS Comp Biol. 2011;7:e1002195.
Tamura K, Stecher G, Kumar S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol Biol Evol. 2021;38:3022–27.
datasets: www.ncbi.nlm.nih.gov/datasets
Entrez Help [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2005-. Entrez Help. 2006 Jan 20 [Updated 2016 May 31]. Available from: https://www.ncbi.nlm.nih.gov/books/NBK3836/
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504.
Acknowledgements
This work was partly supported by Hokkaido University, Global Facility Center (GFC), Pharma Science Open Unit (PSOU), funded by MEXT under “Support Program for Implementation of New Equipment Sharing System”, Global Station for Biosurfaces and Drug Discovery, a project of Global Institution for Collaborative Research and Education in Hokkaido University, the Asahi Glass Foundation, the Naito Foundation, the Uehara Memorial Foundation, the Sumitomo Foundation−Grant for Basic Science Research Projects, Daiichi Sankyo Foundation of Life Science, Japan Foundation for Applied Enzymology, TERUMO Life Science Foundation, the Japan Agency for Medical Research and Development (AMED Grant Numbers JP23ak0101163, JP23ama121039, JP23gm1610007), Grants-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology (MEXT), the Japan Science and Technology Agency (JST Grant Numbers ACT-X JPMJAX201F and A-STEP JPMJTR20US), JSPS KAKENHI (Grant numbers JP16H06448, JP18H02581, JP19K16390, JP21H02635, JP22K15302, JP22H05128 and JP23K17410).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Matsuda, K., Maruyama, H., Imachi, K. et al. Actinobacterial chalkophores: the biosynthesis of hazimycins. J Antibiot 77, 228–237 (2024). https://doi.org/10.1038/s41429-024-00706-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41429-024-00706-6