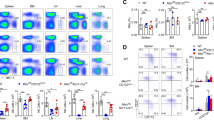
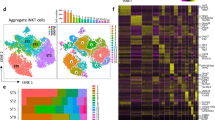
Abstract
Eomesodermin (Eomes) is a critical factor in the development of natural killer (NK) cells, but its precise role in temporal and spatial coordination during this process remains unclear. Our study revealed that Eomes plays distinct roles during the early and late stages of NK cell development. Specifically, the early deletion of Eomes via the CD122-Cre transgene resulted in significant blockade at the progenitor stage due to the downregulation of KLF2, another important transcription factor. ChIP-seq revealed direct binding of Eomes to the conserved noncoding sequence (CNS) of Klf2. Utilizing the CHimeric IMmune Editing (CHIME) technique, we found that deletion of the CNS region of Klf2 via CRISPRi led to a reduction in the NK cell population and developmental arrest. Moreover, constitutive activation of this specific CNS region through CRISPRa significantly reversed the severe defects in NK cell development caused by Eomes deficiency. Conversely, Ncr1-Cre-mediated terminal deletion of Eomes expedited the transition of NK cell subsets from the CD27+CD11b+ phenotype to the CD27−CD11b+ phenotype. Late-stage deficiency of Eomes led to a significant increase in T-bet expression, which subsequently increased the expression of the transcription factor Zeb2. Genetic deletion of one allele of Tbx21, encoding T-bet, effectively reversed the aberrant differentiation of Eomes-deficient NK cells. In summary, we utilized two innovative genetic models to elucidate the intricate mechanisms underlying Eomes-mediated NK cell commitment and differentiation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding author upon request.
References
Vivier E, Raulet DH, Moretta A, Caligiuri MA, Zitvogel L, Lanier LL, et al. Innate or adaptive immunity? The example of natural killer cells. Science. 2011;331:44–9.
Lanier LL. Evolutionary struggles between NK cells and viruses. Nat Rev Immunol. 2008;8:259–68.
Cooper MA, Colonna M, Yokoyama WM. Hidden talents of natural killers: NK cells in innate and adaptive immunity. EMBO Rep. 2009;10:1103–10.
Hayakawa Y, Smyth MJ. CD27 dissects mature NK cells into two subsets with distinct responsiveness and migratory capacity. J Immunol. 2006;176:1517–24.
Chiossone L, Chaix J, Fuseri N, Roth C, Vivier E, Walzer T. Maturation of mouse NK cells is a 4-stage developmental program. Blood. 2009;113:5488–96.
Kee BL, Morman RE, Sun M. Transcriptional regulation of natural killer cell development and maturation. Adv Immunol. 2020;146:1–28.
Geiger TL, Sun JC. Development and maturation of natural killer cells. Curr Opin Immunol. 2016;39:82–9.
Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, Glimcher LH. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 2000;100:655–69.
Zhang J, Le Gras S, Pouxvielh K, Faure F, Fallone L, Kern N, et al. Sequential actions of EOMES and T-BET promote stepwise maturation of natural killer cells. Nat Commun. 2021;12:5446.
Intlekofer AM, Takemoto N, Wherry EJ, Longworth SA, Northrup JT, Palanivel VR, et al. Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat Immunol. 2005;6:1236–44.
Cruz-Guilloty F, Pipkin ME, Djuretic IM, Levanon D, Lotem J, Lichtenheld MG, et al. Runx3 and T-box proteins cooperate to establish the transcriptional program of effector CTLs. J Exp Med. 2009;206:51–9.
Pipkin ME, Sacks JA, Cruz-Guilloty F, Lichtenheld MG, Bevan MJ, Rao A. Interleukin-2 and inflammation induce distinct transcriptional programs that promote the differentiation of effector cytolytic T cells. Immunity. 2010;32:79–90.
Rabacal W, Pabbisetty SK, Hoek KL, Cendron D, Guo Y, Maseda D, et al. Transcription factor KLF2 regulates homeostatic NK cell proliferation and survival. Proc Natl Acad Sci USA. 2016;113:5370–5.
Hart GT, Wang X, Hogquist KA, Jameson SC. Kruppel-like factor 2 (KLF2) regulates B-cell reactivity, subset differentiation, and trafficking molecule expression. Proc Natl Acad Sci USA. 2011;108:716–21.
Hoek KL, Gordy LE, Collins PL, Parekh VV, Aune TM, Joyce S, et al. Follicular B cell trafficking within the spleen actively restricts humoral immune responses. Immunity. 2010;33:254–65.
Winkelmann R, Sandrock L, Porstner M, Roth E, Mathews M, Hobeika E, et al. B cell homeostasis and plasma cell homing controlled by Kruppel-like factor 2. Proc Natl Acad Sci USA. 2011;108:710–5.
Carlson CM, Endrizzi BT, Wu J, Ding X, Weinreich MA, Walsh ER, et al. Kruppel-like factor 2 regulates thymocyte and T-cell migration. Nature. 2006;442:299–302.
Sebzda E, Zou Z, Lee JS, Wang T, Kahn ML. Transcription factor KLF2 regulates the migration of naive T cells by restricting chemokine receptor expression patterns. Nat Immunol. 2008;9:292–300.
LaFleur MW, Nguyen TH, Coxe MA, Yates KB, Trombley JD, Weiss SA, et al. A CRISPR-Cas9 delivery system for in vivo screening of genes in the immune system. Nat Commun. 2019;10:1668.
Zhao H, Liu Y, Wang L, Jin G, Zhao X, Xu J, et al. Genome-wide fitness gene identification reveals Roquin as a potent suppressor of CD8 T cell expansion and anti-tumor immunity. Cell Rep. 2021;37:110083.
Zhou H, Liu J, Zhou C, Gao N, Rao Z, Li H, et al. In vivo simultaneous transcriptional activation of multiple genes in the brain using CRISPR-dCas9-activator transgenic mice. Nat Neurosci. 2018;21:440–6.
Gordon SM, Chaix J, Rupp LJ, Wu J, Madera S, Sun JC, et al. The transcription factors T-bet and Eomes control key checkpoints of natural killer cell maturation. Immunity. 2012;36:55–67.
Daussy C, Faure F, Mayol K, Viel S, Gasteiger G, Charrier E, et al. T-bet and Eomes instruct the development of two distinct natural killer cell lineages in the liver and in the bone marrow. J Exp Med. 2014;211:563–77.
Yang Y, Xu J, Niu Y, Bromberg JS, Ding Y. T-bet and eomesodermin play critical roles in directing T cell differentiation to Th1 versus Th17. J Immunol. 2008;181:8700–10.
Suto A, Wurster AL, Reiner SL, Grusby MJ. IL-21 inhibits IFN-gamma production in developing Th1 cells through the repression of Eomesodermin expression. J Immunol. 2006;177:3721–7.
McLane LM, Banerjee PP, Cosma GL, Makedonas G, Wherry EJ, Orange JS, et al. Differential localization of T-bet and Eomes in CD8 T cell memory populations. J Immunol. 2013;190:3207–15.
Pikovskaya O, Chaix J, Rothman NJ, Collins A, Chen YH, Scipioni AM, et al. Cutting edge: eomesodermin is sufficient to direct type 1 innate lymphocyte development into the conventional NK lineage. J Immunol. 2016;196:1449–54.
McLane LM, Ngiow SF, Chen Z, Attanasio J, Manne S, Ruthel G, et al. Role of nuclear localization in the regulation and function of T-bet and Eomes in exhausted CD8 T cells. Cell Rep. 2021;35:109120.
Wagner JA, Wong P, Schappe T, Berrien-Elliott MM, Cubitt C, Jaeger N, et al. Stage-specific requirement for eomes in mature Nk cell homeostasis and cytotoxicity. Cell Rep. 2020;31:107720.
van Helden MJ, Goossens S, Daussy C, Mathieu AL, Faure F, Marçais A, et al. Terminal NK cell maturation is controlled by concerted actions of T-bet and Zeb2 and is essential for melanoma rejection. J Exp Med. 2015;212:2015–25.
Omilusik KD, Best JA, Yu B, Goossens S, Weidemann A, Nguyen JV, et al. Transcriptional repressor ZEB2 promotes terminal differentiation of CD8+ effector and memory T cell populations during infection. J Exp Med. 2015;212:2027–39.
Jacobsen JA, Bartom ET, Sigvardsson M, Kee BL. Ezh2 represses transcription of innate lymphoid genes in B lymphocyte progenitors and maintains the B-2 cell fate. J Immunol. 2020;204:1760–9.
Schmidl C, Rendeiro AF, Sheffield NC, Bock C. ChIPmentation: fast, robust, low-input ChIP-seq for histones and transcription factors. Nat Methods. 2015;12:963–5.
He J, Zhao J, Quan Y, Hou X, Yang M, Dong Z. Full activation of kinase protein kinase B by phosphoinositide-dependent protein kinase-1 and mammalian target of rapamycin complex 2 is required for early natural killer cell development and survival. Front Immunol. 2020;11:617404.
Acknowledgements
The research reported in this publication was supported by the Natural Science Foundation of China (32330034, 31830027, 31821003, 82271754 and 82071737), the National Key Research & Developmental Program of China (2022YFF0710602), the Excellent Research and Innovation Team in Anhui Province’s Universities (2023AH010085), and the China Postdoctoral Science Foundation (2020M670296 and 2021T140372).
Author information
Authors and Affiliations
Contributions
JH designed and conducted the experiments, analyzed the data, and wrote the manuscript. DC and WX performed most of the bioinformatics analyses. XH and YQ performed the flow cytometry experiments. MY and ZD designed the study, supervised the research, and revised the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests. ZD is an editorial board member of Cellular & Molecular Immunology, but he has not been involved in the peer review of or decision-making regarding the article.
Ethics
The animal study was reviewed and approved by Tsinghua University.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
He, J., Chen, D., Xiong, W. et al. Eomesodermin spatiotemporally orchestrates the early and late stages of NK cell development by targeting KLF2 and T-bet, respectively. Cell Mol Immunol (2024). https://doi.org/10.1038/s41423-024-01164-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41423-024-01164-8