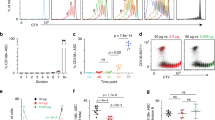
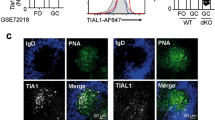
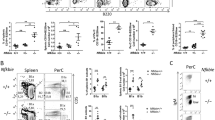
Abstract
Upon recognition of foreign antigens, naïve B cells undergo rapid activation, growth, and proliferation. How B-cell growth and proliferation are coupled with activation remains poorly understood. Combining CRISPR/Cas9-mediated functional analysis and mouse genetics approaches, we found that Dhx33, an activation-induced RNA helicase, plays a critical role in coupling B-cell activation with growth and proliferation. Mutant mice with B-cell-specific deletion of Dhx33 exhibited impaired B-cell development, germinal center reactions, plasma cell differentiation, and antibody production. Dhx33-deficient B cells appeared normal in the steady state and early stage of activation but were retarded in growth and proliferation. Mechanistically, Dhx33 played an indispensable role in activation-induced upregulation of ribosomal DNA (rDNA) transcription. In the absence of Dhx33, activated B cells were compromised in their ability to ramp up 47S ribosomal RNA (rRNA) production and ribosome biogenesis, resulting in nucleolar stress, p53 accumulation, and cellular death. Our findings demonstrate an essential role for Dhx33 in coupling B-cell activation with growth and proliferation and suggest that Dhx33 inhibition is a potential therapy for lymphoma and antibody-mediated autoimmune diseases.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The RNA-seq data generated in this study have been deposited in the NCBI Sequence Read Archive (SRA) under accession number PRJNA850669.
References
Rajewsky K. Clonal selection and learning in the antibody system. Nature. 1996;381:751–8.
Kouzine F, Wojtowicz D, Yamane A, Resch W, Kieffer-Kwon K-R, Bandle R, et al. Global regulation of promoter melting in naive lymphocytes. Cell. 2013;153:988–99.
Wolf T, Jin W, Zoppi G, Vogel IA, Akhmedov M, Bleck CKE, et al. Dynamics in protein translation sustaining T cell preparedness. Nat Immunol. 2020;21:927–37.
Burnet FM. A modification of Jerne’s theory of antibody production using the concept of clonal selection. CA Cancer J Clin. 1976;26:119–21.
Russell J, Zomerdijk JC. RNA-polymerase-I-directed rDNA transcription, life and works. Trends Biochem Sci. 2005;30:87–96.
Bourgeois CF, Mortreux F, Auboeuf D. The multiple functions of RNA helicases as drivers and regulators of gene expression. Nat Rev Mol Cell Bio. 2016;17:426–38.
Takuya N, Kei H, Tatsuya M, Moeko M, Sho M, Ikuo S, et al. In-vitro derived germinal centre B cells differentially generate memory B or plasma cells in vivo. Nat Commun. 2011;2:465.
Hobeika E, Thiemann S, Storch B, Jumaa H, Nielsen PJ, Pelanda R, et al. Testing gene function early in the B cell lineage in mb1-cre mice. Proc Natl Acad Sci USA. 2006;103:13789–94.
Schmidt-Supprian M, Rajewsky K. Vagaries of conditional gene targeting. Nat Immunol. 2007;8:665–8.
Timm W, Jane S, Wiebke W, Tristan W, Van TC, Klaus R, et al. A novel allele for inducible Cre expression in germinal center B cells. Eur J Immunol. 2019;49:192–4.
Zhang Y, Forys JT, Miceli AP. Identification of DHX33 as a mediator of rRNA synthesis and cell growth. Mol Cell Biol. 2011;31:4676–91.
Zhang Y, Lu H. Signaling to p53: Ribosomal proteins find their way. Cancer Cell. 2009;16:369–77.
Su D, Yuan B, Su C, Zhang Y. A 54-kDa short variant of DHX33 functions in regulating mRNA translation. J Cell Physiol. 2019;234:15308–19.
Kaya-Okur HS, Steven JW, Christine AC, Erica SP, Terri DB, Jorja GH, et al. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat Commun. 2019;10:1930.
Kumari P, Tarighi S, Braun T, Ianni A. SIRT7 acts as a guardian of cellular integrity by controlling nucleolar and extra-nucleolar functions. Genes (Basel). 2021;12:1361.
Langst G, Becker PB, Grummt I. TTF-I determines the chromatin architecture of the active rDNA promoter. EMBO J. 1998;17:3135–45.
Grummt I, Langst G. Epigenetic control of RNA polymerase I transcription in mammalian cells. Bba-Gene Regul Mech. 1829;393-404:2013.
Stefanovsky V, Langlois F, Gagnon-Kugler T, Rothblum LI, Moss T. Growth factor signaling regulates elongation of RNA polymerase I transcription in mammals via UBF phosphorylation and r-chromatin remodeling. Mol Cell. 2006;21:629–39.
Wang X, Ge W, Zhang Y. Recombinant DHX33 protein possesses dual DNA/RNA helicase activity. Biochemistry. 2019;58:250–8.
Drygin D, Rice WG, Grummt I. The RNA polymerase I transcription machinery: An emerging target for the treatment of cancer. Annu Rev Pharm Toxicol. 2010;50:131–56.
Bursac S, Prodan Y, Pullen N, Bartek J, Volarevic S. Dysregulated ribosome biogenesis reveals therapeutic liabilities in Cancer. Trends Cancer. 2021;7:57–76.
Fu J, Liu Y, Wang X, Yuan B, Zhang Y. Role of DHX33 in c-Myc-induced cancers. Carcinogenesis. 2017;38:649–60.
Wang H, Yu J, Wang X, Zhang Y. The RNA helicase DHX33 is required for cancer cell proliferation in human glioblastoma and confers resistance to PI3K/mTOR inhibition. Cell Signal. 2019;54:170–8.
Tian Q-H, Zhang M-F, Luo R-G, Fu J, He C, Hu G. et al. DHX33 expression is increased in hepatocellular carcinoma and indicates poor prognosis. Biochem Bioph Res Co. 2016;473:1163–9.
Zhu Y, Du Y, Zhang Y. DHX33 promotes colon cancer development downstream of Wnt signaling. Gene. 2020;735:144402.
Poortinga G, Katherine MH, Hayley S, Carl RW, Anna J, Kerith S, et al. MAD1 and c-MYC regulate UBF and rDNA transcription during granulocyte differentiation. EMBO J. 2004;23:3325–35.
Wang X, Weimin F, Cheng P, Shiyun C, Hongbin J, Hanbing Z, et al. Targeting RNA helicase DHX33 blocks Ras-driven lung tumorigenesis in vivo. Cancer Sci. 2020;111:3564–75.
Wang J, Feng W, Yuan Z, Weber JD, Zhang Y. DHX33 Interacts with AP-2beta to regulate Bcl-2 gene expression and promote cancer cell survival. Mol Cell Biol. 2019;39:e00017-19.
Zhang Y, Saporita AJ, Weber JD. P19ARF and RasV(1)(2) offer opposing regulation of DHX33 translation to dictate tumor cell fate. Mol Cell Biol. 2013;33:1594–607.
Sloan KE, Bohnsack MT, Watkins NJ. The 5S RNP couples p53 homeostasis to ribosome biogenesis and nucleolar stress. Cell Rep. 2013;5:237–47.
Bursac S, Maja CB, Martin P, Ines O, Lior G, Yan Z, et al. Mutual protection of ribosomal proteins L5 and L11 from degradation is essential for p53 activation upon ribosomal biogenesis stress. Proc Natl Acad Sci USA. 2012;109:20467–72.
Donati G, Peddigari S, Mercer CA, Thomas G. 5S ribosomal RNA is an essential component of a nascent ribosomal precursor complex that regulates the Hdm2-p53 checkpoint. Cell Rep. 2013;4:87–98.
Wade M, Li YC, Wahl GM. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat Rev Cancer. 2013;13:83–96.
Martins CP, Brown-Swigart L, Evan GI. Modeling the therapeutic efficacy of p53 restoration in tumors. Cell. 2006;127:1323–34.
Ventura A, David GK, Margaret EM, David AT, Jan G, Laura L, et al. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445:661–5.
Xue W, Lars Z, Cornelius M, Ross AD, Eva H, Valery K, et al. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007;445:656–60.
Platt RJ, Sidi C, Yang Z, Michael JY, Lukasz S, Hannah RK, et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159:440–55.
Rickert RC, Roes J, Rajewsky K. B lymphocyte-specific, Cre-mediated mutagenesis in mice. Nucleic Acids Res. 1997;25:1317–8.
Casola S, Giorgio C, Nathalie U, Sergei BK, Jane S, Zhenyue H, et al. Tracking germinal center B cells expressing germ-line immunoglobulin gamma1 transcripts by conditional gene targeting. Proc Natl Acad Sci USA. 2006;103:7396–401.
Seung GK, Wen-H L, Peiwen L, Hyun YJ, Hyung WL, Jovan S. MicroRNAs of the miR-17~92 family are critical regulators of TFH differentiation. Nat Immunol. 2013;14:849–57.
Yuki H, Takao K, Hiroyuki K, Changshan W, Atsushi I, Keiji K. Downregulation of rRNA transcription triggers cell differentiation. PLoS One. 2014;9:e98586.
Subramanian A, Aravind S, Pablo T, Vamsi KM, Sayan M, Benjamin LE, Michael AG, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50.
Hyun YJ, Hiroyo O, Pengda C, Chao Y, Xiaojuan Z, Seung GK, et al. Differential Sensitivity of Target Genes to Translational Repression by miR-17~92. Plos Genet. 2017;13:e1006623.
Acknowledgements
We thank all members of the CX, W-HL, and Kairui Mao labs (Xiamen University) for technical assistance. This study was supported by the National Natural Science Foundation of China (31570882, 31770950 and 32070877 to W.-H. L, and 81961138008 to CX), the Fundamental Research Funds for the Central Universities of China-Xiamen University (20720170064 to CX), and the Sanofi Institute for Biomedical Research (SIBR).
Author information
Authors and Affiliations
Contributions
CX, HYJ, and PC conceived this project. XH and JZ designed and performed most of the experiments, analyzed data, and generated figures; AA and PH performed some experiments and provided technical assistance; XL performed RNA sequencing analysis; PC, JX, YD, YL, and LL contributed to the experiments; CX and W-HL supervised the project. XH, JZ, W-HL, and CX wrote the manuscript with contributions from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
He, X., Zhao, J., Adilijiang, A. et al. Dhx33 promotes B-cell growth and proliferation by controlling activation-induced rRNA upregulation. Cell Mol Immunol 20, 277–291 (2023). https://doi.org/10.1038/s41423-022-00972-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41423-022-00972-0