Abstract
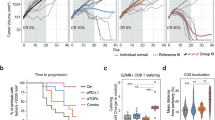
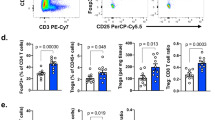
Blockade of programmed death-1 (PD-1) reinvigorates exhausted CD8+ T cells, resulting in tumor regression in cancer patients. Recently, reinvigoration of exhausted CD8+ T cells following PD-1 blockade was shown to be CD28-dependent in mouse models. Herein, we examined the role of CD28 in anti-PD-1 antibody-induced human T cell reinvigoration using tumor-infiltrating CD8+ T cells (CD8+ TILs) obtained from non-small-cell lung cancer patients. Single-cell analysis demonstrated a distinct expression pattern of CD28 between mouse and human CD8+ TILs. Furthermore, we found that human CD28+CD8+ but not CD28–CD8+ TILs responded to PD-1 blockade irrespective of B7/CD28 blockade, indicating that CD28 costimulation in human CD8+ TILs is dispensable for PD-1 blockade-induced reinvigoration and that loss of CD28 expression serves as a marker of anti-PD-1 antibody-unresponsive CD8+ TILs. Transcriptionally and phenotypically, PD-1 blockade-unresponsive human CD28–PD-1+CD8+ TILs exhibited characteristics of terminally exhausted CD8+ T cells with low TCF1 expression. Notably, CD28–PD-1+CD8+ TILs had preserved machinery to respond to IL-15, and IL-15 treatment enhanced the proliferation of CD28–PD-1+CD8+ TILs as well as CD28+PD-1+CD8+ TILs. Taken together, these results show that loss of CD28 expression is a marker of PD-1 blockade-unresponsive human CD8+ TILs with a TCF1– signature and provide mechanistic insights into combining IL-15 with anti-PD-1 antibodies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Ribas, A. & Wolchok, J. D. Cancer immunotherapy using checkpoint blockade. Science. 359, 1350–1355 (2018).
Sharma, P., Hu-Lieskovan, S., Wargo, J. A. & Ribas, A. Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell. 168, 707–723 (2017).
Tang, J. et al. Trial watch: the clinical trial landscape for PD1/PDL1 immune checkpoint inhibitors. Nat. Rev. Drug Discov. 17, 854–855 (2018).
Tumeh, P. C. et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 515, 568–571 (2014).
Huang, A. C. et al. T-cell invigoration to tumour burden ratio associated with anti-PD-1 response. Nature. 545, 60–65 (2017).
Kim, K. H. et al. The first-week proliferative response of peripheral blood PD-1(+)CD8(+) T cells predicts the response to anti-PD-1 therapy in solid tumors. Clin Cancer Res. 25, 2144–2154 (2019).
Blackburn, S. D., Shin, H., Freeman, G. J. & Wherry, E. J. Selective expansion of a subset of exhausted CD8 T cells by alphaPD-L1 blockade. Proc. Natl Acad Sci USA. 105, 15016–15021 (2008).
Paley, M. A. et al. Progenitor and terminal subsets of CD8+ T cells cooperate to contain chronic viral infection. Science. 338, 1220–1225 (2012).
Utzschneider, D. T. et al. T Cell Factor 1-expressing memory-like CD8(+) T cells sustain the immune response to chronic viral infections. Immunity. 45, 415–427 (2016).
Im, S. J. et al. Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature. 537, 417–421 (2016).
He, R. et al. Follicular CXCR5- expressing CD8(+) T cells curtail chronic viral infection. Nature. 537, 412–428 (2016).
Wu T. et al. The TCF1-Bcl6 axis counteracts type I interferon to repress exhaustion and maintain T cell stemness. Sci Immunol. 23, eaai8593 (2016).
Siddiqui, I. et al. Intratumoral Tcf1(+)PD-1(+)CD8(+) T cells with stem-like properties promote tumor control in response to vaccination and checkpoint blockade immunotherapy. Immunity. 50, 195–211 (2019). e110.
Miller, B. C. et al. Subsets of exhausted CD8(+) T cells differentially mediate tumor control and respond to checkpoint blockade. Nat Immunol. 20, 326–336 (2019).
Hui, E. et al. T cell costimulatory receptor CD28 is a primary target for PD-1-mediated inhibition. Science. 355, 1428–1433 (2017).
Kamphorst, A. O. et al. Rescue of exhausted CD8 T cells by PD-1-targeted therapies is CD28-dependent. Science. 355, 1423–1427 (2017).
Guo, X. et al. Global characterization of T cells in non-small-cell lung cancer by single-cell sequencing. Nat Med. 24, 978–985 (2018).
Gubin, M. M. et al. High-dimensional analysis delineates myeloid and lymphoid compartment remodeling during successful immune-checkpoint cancer therapy. Cell. 175, 1014–1030 (2018). e1019.
Huang, A. C. et al. A single dose of neoadjuvant PD-1 blockade predicts clinical outcomes in resectable melanoma. Nat Med. 25, 454–461 (2019).
Philip, M. et al. Chromatin states define tumour-specific T cell dysfunction and reprogramming. Nature. 545, 452–456 (2017).
Thommen, D. S. et al. A transcriptionally and functionally distinct PD-1(+) CD8(+) T cell pool with predictive potential in non-small-cell lung cancer treated with PD-1 blockade. Nat Med. 24, 994–1004 (2018).
Zheng, C. et al. Landscape of infiltrating T cells in liver cancer revealed by single-cell sequencing. Cell. 169, 1342–1356 (2017). e1316.
Tirosh, I. et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 352, 189–196 (2016).
Quigley, M. et al. Transcriptional analysis of HIV-specific CD8+ T cells shows that PD-1 inhibits T cell function by upregulating BATF. Nat Med. 16, 1147–1151 (2010).
Gupta, P. K. et al. CD39 expression identifies terminally exhausted CD8+ T cells. PLoS Pathog. 11, e1005177 (2015).
Wherry, E. J. et al. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity. 27, 670–684 (2007).
Bezman, N. A. et al. Molecular definition of the identity and activation of natural killer cells. Nat Immunol. 13, 1000–1009 (2012).
Safford, M. et al. Egr-2 and Egr-3 are negative regulators of T cell activation. Nat Immunol. 6, 472–480 (2005).
Fridman, A. L. & Tainsky, M. A. Critical pathways in cellular senescence and immortalization revealed by gene expression profiling. Oncogene. 27, 5975–5987 (2008).
Weng, N. P., Akbar, A. N. & Goronzy, J. CD28(-) T cells: their role in the age-associated decline of immune function. Trends Immunol. 30, 306–312 (2009).
Hugo, W. et al. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell. 165, 35–44 (2016).
Wrangle, J. M. et al. ALT-803, an IL-15 superagonist, in combination with nivolumab in patients with metastatic non-small cell lung cancer: a non-randomised, open-label, phase 1b trial. Lancet Oncol. 19, 694–704 (2018).
Kurtulus, S. et al. Checkpoint blockade immunotherapy induces dynamic changes in PD-1(-)CD8(+) tumor-infiltrating T cells. Immunity. 50, 181–194 (2019). e186.
Sanchez-Paulete, A. R. et al. Cancer Immunotherapy with immunomodulatory anti-CD137 and anti-PD-1 monoclonal antibodies requires BATF3-dependent dendritic cells. Cancer Discov. 6, 71–79 (2016).
Snell, L. M. et al. CD8(+) T cell priming in established chronic viral infection preferentially directs differentiation of memory-like cells for sustained immunity. Immunity. 49, 678–694 (2018). e675.
Chen, Z. et al. TCF-1-centered transcriptional network drives an effector versus exhausted CD8 T cell-fate decision. Immunity. 51, 840–855 (2019). e845.
Sade-Feldman, M. et al. Defining T cell states associated with response to checkpoint immunotherapy in melanoma. Cell. 175, 998–1013 (2018). e1020.
Gattinoni, L. et al. Wnt signaling arrests effector T cell differentiation and generates CD8+ memory stem cells. Nat Med. 15, 808–813 (2009).
Jeannet, G. et al. Essential role of the Wnt pathway effector Tcf-1 for the establishment of functional CD8 T cell memory. Proc Natl Acad Sci USA. 107, 9777–9782 (2010).
Sabatino, M. et al. Generation of clinical-grade CD19-specific CAR-modified CD8+ memory stem cells for the treatment of human B-cell malignancies. Blood. 128, 519–528 (2016).
Zhan, T., Rindtorff, N. & Boutros, M. Wnt signaling in cancer. Oncogene. 36, 1461–1473 (2017).
Ruiz de Galarreta M., et al. beta-catenin activation promotes immune escape and resistance to anti-PD-1 therapy in hepatocellular carcinoma. Cancer Discov. 2019; https://doi.org/10.1158/2159-8290.CD-19-0074.
Nsengimana, J. et al. beta-Catenin-mediated immune evasion pathway frequently operates in primary cutaneous melanomas. J Clin Investig. 128, 2048–2063 (2018).
Xue, J. et al. Intrinsic beta-catenin signaling suppresses CD8(+) T-cell infiltration in colorectal cancer. Biomed Pharmacother. 115, 108921 (2019).
Jabri, B. & Abadie, V. IL-15 functions as a danger signal to regulate tissue-resident T cells and tissue destruction. Nat Rev Immunol. 15, 771–783 (2015).
Waldmann, T. A. The biology of interleukin-2 and interleukin-15: implications for cancer therapy and vaccine design. Nat Rev Immunol. 6, 595–601 (2006).
Shin, H., Blackburn, S. D., Blattman, J. N. & Wherry, E. J. Viral antigen and extensive division maintain virus-specific CD8 T cells during chronic infection. J Exp Med. 204, 941–949 (2007).
Liu, R. B. et al. IL-15 in tumor microenvironment causes rejection of large established tumors by T cells in a noncognate T cell receptor-dependent manner. Proc Natl Acad Sci USA. 110, 8158–8163 (2013).
Mathios, D. et al. Therapeutic administration of IL-15 superagonist complex ALT-803 leads to long-term survival and durable antitumor immune response in a murine glioblastoma model. Int J Cancer. 138, 187–194 (2016).
Conlon K. et al. Abstract CT082: phase (Ph) I/Ib study of NIZ985 with and without spartalizumab (PDR001) in patients (pts) with metastatic/unresectable solid tumors. Cancer Res. 79, CT082 (2019).
Simoni, Y. et al. Bystander CD8(+) T cells are abundant and phenotypically distinct in human tumour infiltrates. Nature. 557, 575–579 (2018).
Duhen, T. et al. Co-expression of CD39 and CD103 identifies tumor-reactive CD8 T cells in human solid tumors. Nat Commun. 9, 2724 (2018).
Scheper, W. et al. Low and variable tumor reactivity of the intratumoral TCR repertoire in human cancers. Nat Med. 25, 89–94 (2019).
McLane, L. M., Abdel-Hakeem, M. S. & Wherry, E. J. CD8 T cell exhaustion during chronic viral infection and cancer. Annu Rev Immunol. 37, 457–495 (2019).
Kim, H. D. et al. Association between expression level of PD1 by tumor-infiltrating CD8(+) T cells and features of hepatocellular carcinoma. Gastroenterology. 155, 1936–1950 (2018). e1917.
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9, 357–359 (2012).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 26, 841–842 (2010).
Hanzelmann, S., Castelo, R. & Guinney, J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 14, 7 (2013).
Newman, A. M. et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 12, 453–457 (2015).
Satija, R., Farrell, J. A., Gennert, D., Schier, A. F. & Regev, A. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol. 33, 495–502 (2015).
Author information
Authors and Affiliations
Contributions
KHK, HKK, M-JA, and E-CS contributed to the conceptual design of the study. HKK, M-JA, JK, and BMK collected human samples. KHK, HL, and HK processed patients’ samples. KHK and E-CS designed the experiments. KHK, H-DK, CGK, HL, JWH, SJC were involved in data acquisition. KHK, HKK, H-DK, CGK, HL, JWH, SJC, S-HP, M-JA, and E-CS were involved in data analysis and interpretation. KHK performed statistical analysis. KHK, HKK, M-JA, and E-CS drafted the manuscript. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Supplementary information
Rights and permissions
About this article
Cite this article
Kim, K.H., Kim, H.K., Kim, HD. et al. PD-1 blockade-unresponsive human tumor-infiltrating CD8+ T cells are marked by loss of CD28 expression and rescued by IL-15. Cell Mol Immunol 18, 385–397 (2021). https://doi.org/10.1038/s41423-020-0427-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41423-020-0427-6
Keywords
This article is cited by
-
Construction of an immune-related risk score signature for gastric cancer based on multi-omics data
Scientific Reports (2024)
-
CD8+ T cell metabolic flexibility elicited by CD28-ARS2 axis-driven alternative splicing of PKM supports antitumor immunity
Cellular & Molecular Immunology (2024)
-
CD28/PD1 co-expression: dual impact on CD8+ T cells in peripheral blood and tumor tissue, and its significance in NSCLC patients' survival and ICB response
Journal of Experimental & Clinical Cancer Research (2023)
-
Comparison of the tumor immune microenvironment and checkpoint blockade biomarkers between stage III and IV non-small cell lung cancer
Cancer Immunology, Immunotherapy (2023)
-
Novel targets for immunotherapy associated with exhausted CD8 + T cells in cancer
Journal of Cancer Research and Clinical Oncology (2023)