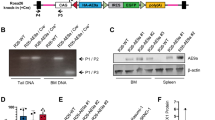
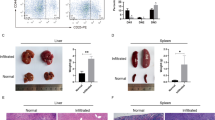
Abstract
Tcf-1 (encoded by Tcf7) not only plays critical roles in promoting T cell development and differentiation but also has been identified as a tumor suppressor involved in preventing T cell malignancy. However, the comprehensive mechanisms of Tcf-1 involved in T cell transformation remain poorly understood. In this study, Tcf7fl/fl mice were crossed with Vav-cre, Lck-cre, or Cd4-cre mice to delete Tcf-1 conditionally at the beginning of the HSC, DN2–DN3, or DP stage, respectively. The defective T cell development phenotypes became gradually less severe as the deletion stage became more advanced in distinct mouse models. Interestingly, consistent with Tcf7−/− mice, Tcf7fl/flVav-cre mice developed aggressive T cell lymphoma within 45 weeks, but no tumors were generated in Tcf7fl/flLck-cre or Tcf7fl/flCd4-cre mice. Single-cell RNA-seq (ScRNA-seq) indicated that ablation of Tcf-1 at distinct phases can subdivide DN1 cells into three clusters (C1, C2, and C3) and DN2–DN3 cells into three clusters (C4, C5, and C6). Moreover, Tcf-1 deficiency redirects bifurcation among divergent cell fates, and clusters C1 and C4 exhibit high potential for leukemic transformation. Mechanistically, we found that Tcf-1 directly binds and mediates chromatin accessibility for both typical T cell regulators and proto-oncogenes, including Myb, Mycn, Runx1, and Lyl1 in the DN1 phase and Lef1, Id2, Dtx1, Fyn, Bcl11b, and Zfp36l2 in the DN2–DN3 phase. The aberrant expression of these genes due to Tcf-1 deficiency in very early T cells contributes to subsequent tumorigenesis. Thus, we demonstrated that Tcf-1 plays stage-specific roles in regulating early thymocyte development and transformation, providing new insights and evidence for clinical trials on T-ALL leukemia.
This is a preview of subscription content, access via your institution
Access options







Similar content being viewed by others
References
Rothenberg, E. V., Moore, J. E. & Yui, M. A. Launching the T-cell-lineage developmental programme. Nat. Rev. Immunol. 8, 9–21 (2008).
Ramond, C. et al. Two waves of distinct hematopoietic progenitor cells colonize the fetal thymus. Nat. Immunol. 15, 27–35 (2014).
Yu, S., Zhao, D. M., Jothi, R. & Xue, H. H. Critical requirement of GABPalpha for normal T cell development. J. Biol. Chem. 285, 10179–10188 (2010).
Rothenberg, E. V., Kueh, H. Y., Yui, M. A. & Zhang, J. A. Hematopoiesis and T-cell specification as a model developmental system. Immunol. Rev. 271, 72–97 (2016).
Vicente, R. et al. Molecular and cellular basis of T cell lineage commitment. Semin. Immunol. 22, 270–275 (2010).
Yui, M. A. & Rothenberg, E. V. Developmental gene networks: a triathlon on the course to T cell identity. Nat. Rev. Immunol. 14, 529–545 (2014).
Georgopoulos, K. Haematopoietic cell-fate decisions, chromatin regulation and ikaros. Nat. Rev. Immunol. 2, 162–174 (2002).
Maillard, I., Fang, T. & Pear, W. S. Regulation of lymphoid development, differentiation, and function by the Notch pathway. Annu. Rev. Immunol. 23, 945–974 (2005).
Johnson, J. L. et al. Lineage-determining transcription factor TCF-1 initiates the epigenetic identity of T cells. Immunity 48, 243–257 e10 (2018).
Weng, A. P. et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science 306, 269–271 (2004).
Aifantis, I., Raetz, E. & Buonamici, S. Molecular pathogenesis of T-cell leukaemia and lymphoma. Nat. Rev. Immunol. 8, 380–390 (2008).
Haydu, J. E. & Ferrando, A. A. Early T-cell precursor acute lymphoblastic leukaemia. Curr. Opin. Hematol. 20, 369–373 (2013).
McCormack, M. P. et al. The Lmo2 oncogene initiates leukemia in mice by inducing thymocyte self-renewal. Science 327, 879–883 (2010).
McCormack, M. P. et al. Requirement for Lyl1 in a model of Lmo2-driven early T-cell precursor ALL. Blood 122, 2093–2103 (2013).
Grabher, C., von Boehmer, H. & Look, A. T. Notch 1 activation in the molecular pathogenesis of T-cell acute lymphoblastic leukaemia. Nat. Rev. Cancer 6, 347–359 (2006).
Yu, S. & Xue, H. H. TCF-1 mediates repression of Notch pathway in T lineage-committed early thymocytes. Blood 121, 4008–4009 (2013).
Yu, S. et al. The TCF-1 and LEF-1 transcription factors have cooperative and opposing roles in T cell development and malignancy. Immunity 37, 813–826 (2012).
Verbeek, S. et al. An HMG-box-containing T-cell factor required for thymocyte differentiation. Nature 374, 70–74 (1995).
Steinke, F. C. et al. TCF-1 and LEF-1 act upstream of Th-POK to promote the CD4(+) T cell fate and interact with Runx3 to silence Cd4 in CD8(+) T cells. Nat. Immunol. 15, 646–56. (2014).
Held, W., Clevers, H. & Grosschedl, R. Redundant functions of TCF-1 and LEF-1 during T and NK cell development, but unique role of TCF-1 for Ly49 NK cell receptor acquisition. Eur. J. Immunol. 33, 1393–1398 (2003).
Schilham, M. W. et al. Critical involvement of Tcf-1 in expansion of thymocytes. J. Immunol. 161, 3984–3991 (1998).
Ioannidis, V., Beermann, F., Clevers, H. & Held, W. The beta-catenin-TCF-1 pathway ensures CD4(+)CD8(+) thymocyte survival. Nat. Immunol. 2, 691–697 (2001).
Goux, D. et al. Cooperating pre-T-cell receptor and TCF-1-dependent signals ensure thymocyte survival. Blood 106, 1726–1733 (2005).
Weber, B. N. et al. A critical role for TCF-1 in T-lineage specification and differentiation. Nature 476, 63–68 (2011).
Germar, K. et al. T-cell factor 1 is a gatekeeper for T-cell specification in response to Notch signaling. Proc. Natl Acad. Sci. USA 108, 20060–20065 (2011).
Zhao, D. M. et al. Constitutive activation of Wnt signaling favors generation of memory CD8 T cells. J. Immunol. 184, 1191–1199 (2010).
Zhou, X. et al. Differentiation and persistence of memory CD8(+) T cells depend on T cell factor 1. Immunity 33, 229–240 (2010).
Choi, Y. S. et al. LEF-1 and TCF-1 orchestrate T(FH) differentiation by regulating differentiation circuits upstream of the transcriptional repressor Bcl6. Nat. Immunol. 16, 980–990 (2015).
Wu, T. et al. TCF1 is required for the T follicular helper cell response to viral infection. Cell Rep. 12, 2099–2110 (2015).
Xu, L. et al. The transcription factor TCF-1 initiates the differentiation of T(FH) cells during acute viral infection. Nat. Immunol. 16, 991–999 (2015).
Tiemessen, M. M. et al. The nuclear effector of Wnt-signaling, Tcf1, functions as a T-cell-specific tumor suppressor for development of lymphomas. PLoS Biol. 10, e1001430 (2012).
de Boer, J. et al. Transgenic mice with hematopoietic and lymphoid specific expression of Cre. Eur. J. Immunol. 33, 314–325 (2003).
Lee, P. P. et al. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity 15, 763–774 (2001).
Tirosh, I. et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 352, 189–196 (2016).
Qiu, X. et al. Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods 14, 979–82. (2017).
Choi, A. et al. RUNX1 is required for oncogenic Myb and Myc enhancer activity in T-cell acute lymphoblastic leukemia. Blood 130, 1722–33. (2017).
Souroullas, G. P., Salmon, J. M., Sablitzky, F., Curtis, D. J. & Goodell, M. A. Adult hematopoietic stem and progenitor cells require either Lyl1 or Scl for survival. Cell Stem Cell 4, 180–186 (2009).
McCormack, M. P. et al. Requirement for Lyl1 in a model of Lmo2-driven early T-cell precursor ALL. Blood 122, 2093–2103 (2013).
Astolfi, A. et al. MYCN is a novel oncogenic target in pediatric T-cell acute lymphoblastic leukemia. Oncotarget 5, 120–130 (2014).
Smith, S. et al. LIM domain only-2 (LMO2) induces T-cell leukemia by two distinct pathways. PLoS ONE 9, e85883 (2014).
Xing, S. et al. Tcf1 and Lef1 transcription factors establish CD8(+) T cell identity through intrinsic HDAC activity. Nat. Immunol. 17, 695–703 (2016).
Georgescu, C. et al. A gene regulatory network armature for T lymphocyte specification. Proc. Natl Acad. Sci. USA 105, 20100–20105 (2008).
Mielke, L. A. et al. TCF-1 limits the formation of Tc17 cells via repression of the MAF-ROR gamma t axis. J. Exp. Med. 216, 1682–99. (2019).
Raghu, D., Xue, H. H. & Mielke, L. A. Control of lymphocyte fate, infection, and tumor immunity by TCF-1. Trends Immunol. 40, 1149–62. (2019).
Held, W., Kunz, B., Lowin-Kropf, B., van de Wetering, M. & Clevers, H. Clonal acquisition of the Ly49A NK cell receptor is dependent on the trans-acting factor TCF-1. Immunity 11, 433–442 (1999).
Mielke, L. A. et al. TCF-1 controls ILC2 and NKp46+RORgammat+ innate lymphocyte differentiation and protection in intestinal inflammation. J. Immunol. 191, 4383–4391 (2013).
Yang, Q. et al. T cell factor 1 is required for group 2 innate lymphoid cell generation. Immunity 38, 694–704 (2013).
Martins, V. C. et al. Cell competition is a tumour suppressor mechanism in the thymus. Nature 509, 465–470 (2014).
Yu, S. et al. GABP controls a critical transcription regulatory module that is essential for maintenance and differentiation of hematopoietic stem/progenitor cells. Blood 117, 2166–2178 (2011).
Hodson, D. J. et al. Deletion of the RNA-binding proteins ZFP36L1 and ZFP36L2 leads to perturbed thymic development and T lymphoblastic leukemia. Nat. Immunol. 11, 717–724 (2010).
Yu, G. et al. TCF-1 deficiency influences the composition of intestinal microbiota and enhances susceptibility to colonic inflammation. Protein Cell 11, 380–386 (2020).
Cui, Z. Z. et al. Over-expression of CD163, CD169, and CD151 is not sufficient to improve the susceptibility to porcine reproductive and respiratory syndrome virus infection in transgenic mice. Sci. Bull. 62, 1634–1636 (2017).
Dong, J. et al. Single-cell RNA-seq analysis unveils a prevalent epithelial/mesenchymal hybrid state during mouse organogenesis. Genome Biol. 19, 31 (2018).
Trapnell, C., Pachter, L. & Salzberg, S. L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 (2009).
Li, L. et al. A far downstream enhancer for murine Bcl11b controls its T-cell specific expression. Blood 122, 902–911 (2013).
Szklarczyk, D. et al. The STRING database in 2017: quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 45, D362–D8. (2017).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Acknowledgements
We thank Mr. Biliang Zhang and Dr. Ziding Zhang at the College of Biological Sciences, China Agricultural University, for their constructive comments and assistance with the computational analysis. We also thank Dr. Bing Zhang at the Laboratory Animal Center at China Agricultural University for his assistance with animal care. This work was supported by the National Key R&D Program of China (2017YFA0104401), the National Natural Science Foundation of China (31630038, 31970831, and 31571522), and the Project for Extramural Scientists of State Key Laboratory of Agrobiotechnology from China Agricultural University (2019SKLAB6-6 & 2019SKLAB6-7).
Author information
Authors and Affiliations
Contributions
S.Y. designed and supervised the experiments. F.W. analyzed the high-throughput data under the supervision of P.J.; single cells were isolated and selected by G.Y. and T.F. F.W., Z.Q., Y.Y., and G.Y. performed the general experiments. T.Z., H.-H.X., Y.Z., L.B., and S.Y. assisted with the general experiments and interpreted the data. S.Y. and F.W. wrote the manuscript with the aid of all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Rights and permissions
About this article
Cite this article
Wang, F., Qi, Z., Yao, Y. et al. Exploring the stage-specific roles of Tcf-1 in T cell development and malignancy at single-cell resolution. Cell Mol Immunol 18, 644–659 (2021). https://doi.org/10.1038/s41423-020-00527-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41423-020-00527-1
Keywords
This article is cited by
-
Chromatin organizer SATB1 controls the cell identity of CD4+ CD8+ double-positive thymocytes by regulating the activity of super-enhancers
Nature Communications (2022)
-
TCF1 in T cell immunity: a broadened frontier
Nature Reviews Immunology (2022)
-
RNA polymerase II pausing factor NELF in CD8+ T cells promotes antitumor immunity
Nature Communications (2022)