Abstract
Gallbladder cancer (GBC) is the most common malignant tumour of the biliary track system. Angiogenesis plays a pivotal role in the development and progression of malignant tumours. miR-143-3p acts as a tumour suppressor in various cancers. Their role in GBC is however less well defined. Here we show that the expression levels of miR-143-3p were decreased in human GBC tissues compared with the non-tumour adjacent tissue (NAT) counterparts and were closely associated with overall survival. We discovered that miR-143-3p was a novel inhibitor of tumour growth and angiogenesis in vivo and in vitro. Our antibody array, ELISA and PLGF rescue analyses indicated that PLGF played an essential role in the antiangiogenic effect of miR-143-3p. Furthermore, we used miRNA target-prediction software and dual-luciferase assays to confirm that integrin α6 (ITGA6) acted as a direct target of miR-143-3p. Our ELISA and western blot analyses confirmed that the expression of PLGF was decreased via the ITGA6/PI3K/AKT pathway. In conclusion, miR-143-3p suppresses tumour angiogenesis and growth of GBC through the ITGA6/PI3K/AKT/PLGF pathways and may be a novel molecular therapeutic target for GBC.
Similar content being viewed by others
Introduction
Gallbladder cancer (GBC) is the most common malignant tumour of the biliary track system1 and the fifth most common gastrointestinal cancer2,3,4. The Surveillance, Epidemiology, and End Results (SEER) programme has shown the incidence of gallbladder carcinoma to be approximately 2.5 cases per 1×105 people1,2,3,4. Although the incidence of GBC is lower than the incidences of other gastrointestinal cancers, such as gastric cancer, the survival rate for GBC is poor due to difficulties with early diagnosis, the frequent occurrence of early metastasis and the high degree of malignancy; the five-year survival rate of GBC is less than 5%5. Surgical resection is the only effective treatment method because GBC is not sensitive to radiotherapy and chemotherapy6. Despite its atypical early symptoms, patients have no opportunity to undergo surgery. Therefore, novel prognostic biomarkers and targeted therapeutics for GBC are necessary7.
Angiogenesis plays a central role in the development and progression of malignant tumours8. Increases in angiogenic factors and reductions in antiangiogenic factors contribute to the formation of new blood vessels9. Vascular endothelial growth factor (VEGF) is widely recognized as a significant angiogenic factor. In addition to VEGF, another group of proteins, including fibroblast growth factor 2 (FGF2), angiopoietin-1 (Ang-1) and platelet-derived growth factor receptor-alpha (PDGFRα), directly participate in the formation of blood capillaries and lymphatic vessels. The VEGF family comprises six secretory glycoproteins, namely, VEGFA, VEGFB, VEGFC, VEGFD, VEGFE and placental growth factor (PLGF)10. Previous studies have reported that PLGF plays pivotal roles in pathological contexts such as cancer11, whereas PLGF actions are redundant in normal physiological processes. PLGF binds to and activates VEGF receptor 1 (Flt-1) and synergizes the effect of VEGF11. Numerous small-molecule inhibitors of VEGF and VEGFR2, including cediranib12, sunitinib13 and vandetanib14, have been approved for therapy. However, significant increases in other angiogenic regulatory factors during cancer development have also been observed following treatment. Thus, a better understanding of the process of angiogenesis is critical to overcoming the side-effects of antiangiogenic approaches and developing a new effective antiangiogenic therapy.
MicroRNAs (miRNAs) are a class of highly conserved small (18–24 nucleotides in length) noncoding RNAs that can inhibit translation or induce mRNA degradation by binding to the 3′-untranslated regions (3’-UTRs) of target genes15. As post-transcriptional regulators, miRNAs play significant roles in physiological and pathological processes, including cell differentiation, organ development, cell proliferation, apoptosis and tumourigenesis16. miRNAs can act as either oncogenes or tumour suppressors by directly or indirectly modulating cancer genes17. Accumulative studies have demonstrated that miRNAs play significant roles in GBC. According to a previous study, miR-143-3p is dramatically downregulated in GBC tissues compared with non-tumour adjacent tissues (NATs)1,18,19. However, the precise molecular mechanism through which miR-143-3p influences the progression of GBC remains unknown.
In this study, we aimed to evaluate the underlying roles and mechanisms of miR-143-3p in gallbladder tumourigenesis. We found that the expression level of miR-143-3p was significantly lower in GBC tissues than in the NAT counterparts. Additionally, we found that miR-143-3p played vital roles in GBC angiogenesis and growth. miR-143-3p inhibited the function of integrin α6 (ITGA6), a direct target of miR-143-3p, which induced tumour angiogenesis through the MAPK and PI3K/AKT pathways and enhanced the expression of PLGF. Here, we first determined the inhibitory role of miR-143-3p in GBC growth and angiogenesis and then demonstrated the potential use of miR-143-3p as a targeted therapy and as a prognostic indicator for patients with GBC.
Results
Downregulation of miR-143-3p is associated with an unfavourable prognosis in GBC patients
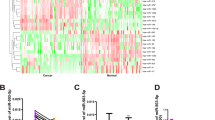
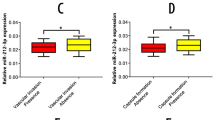
According to our previous microarray results1, miR-143-3p was dramatically downregulated in GBC tissues compared with NATs (Fig. 1a). To validate our previous miRNA profiling data, we evaluated miR-143-3p expression in 49 paired clinical GBC specimens. The expression levels of miR-143-3p were significantly lower in the tumour tissues than in the corresponding adjacent noncancerous tissues (P = 0.0021, Fig. 1b, c). In the survival analysis, 49 GBC patients were divided into two groups (miR-143-3p-low, n = 25; miR-143-3p-high, n = 24) by setting the cut-off value to the median miR-143-3p expression level. The Kaplan−Meier analysis revealed that high miR-143-3p expression was associated with better overall survival compared with lower expression levels (P < 0.001) (Fig. 1d). The mean survival time in the low miR-143-3p expression group was 9.86 months, whereas the mean survival time in the high miR-143-3p expression group was 26.92 months. A clinicopathological association study of the 49 GBCs showed that miR-143-3p was significantly associated with tumour size (P = 0.016) (Table 1) and tumour invasion (P = 0.023) (Table 1).
a A portion of the cluster analysis of the miRNA expression profiles of the GBC tissues and NAT counterparts from our previous microarray results. b Scatterplots of the relative expression levels of miR-143-3p in the GBC tissues and their corresponding NATs. miR-143-3p expression was calculated and expressed as the miR-143-3p/U6 expression ratio (2−ΔCT). P = 0.0021. c Comparison of the miR-143-3p expression levels between the GBC tissues and their corresponding NATs. d Kaplan−Meier overall survival curve of GBC patients based on miR-143-3p expression. P < 0.001
miR-143-3p inhibits GBC cell proliferation and angiogenesis in vitro
We first assessed the expression levels of miR-143-3p in five human GBC cell lines (NOZ, GBC-SD, SGC996, OCUG-1 and EHGB-1). The miR-143-3p expression levels were lowest in NOZ cells and highest in GBC-SD cells among five GBC cell lines (Supplementary Figure S1a). Therefore, we selected NOZ and GBC-SD cells for subsequent studies. To investigate the biological functions of miR-143-3p in GBC angiogenesis, we performed a gain- and loss-of-function analysis using miR-143-3p mimics and inhibitors. The effects of the mimics and inhibitors were examined using qRT-PCR (Supplementary Figure S1b and c). The conditional medium after 48 h of culture was collected and used for human microvascular vein endothelial cell (HMVEC) tube formation and invasion assays to determine the effects of miR-143-3p on angiogenesis in vitro. As a result, the culture medium from the NOZ and GBC-SD cells transfected with the miR-143-3p mimics significantly impaired capillary tube formation and decreased the invasive abilities of HMVECs (Fig. 2a−d left). Conversely, HMVECs formed more branch points in the group of GBC cells transfected with the miR-143-3p inhibitors than in the inhibitor negative control (NC) group. Additionally, higher invasive activity was evident in HMVECs treated with the conditional medium of GBC cells transfected with the miR-143-3p inhibitors (Fig. 2a−d right). CCK-8 assays were performed to assess the role of miR-143-3p in NOZ and GBC-SD cell proliferation. The GBC cells transfected with the miR-143-3p mimics clearly grew more slowly than the mimic NC group. However, the proliferation rate of GBC cells transfected with the miR-143-3p inhibitors was significantly increased relative to the inhibitor NC group (Fig. 2e and Supplementary Figure 1d). These findings suggest that miR-143-3p suppresses GBC cell proliferation and angiogenesis in vitro.
a, b Endothelial tube formation was estimated following incubation of HMVECs with conditioned medium from GBC cells transfected with mimics or inhibitors. The number of branches was quantified (**P < 0.01, ***P < 0.001; Student’s t test). Scale bar, 100 µm. c, d Invasion of HMVECs through the Matrigel chambers after incubation with conditioned medium from miR-143-3p-overexpressing or miR-143-3p-inhibited GBC cells for 48 h. Scale bar, 100 µm. The number of invading cells was calculated and is depicted in the bar graph (**P < 0.01, ***P < 0.001; Student’s t test). e Cell growth rates over 5 days were determined with CCK-8 proliferation assays (**P < 0.01, ***P < 0.001)
miR-143-3p inhibits GBC cell proliferation and angiogenesis in vivo
To confirm these findings in vivo, NOZ cells stably expressing miR-143-3p were subcutaneously implanted into 4-week-old nude mice to investigate the effect of miR-143-3p on tumour growth. The results showed that the volumes and weights of xenografted tumours were smaller and lower, respectively, in the miR-143-3p overexpression group (Lv-miR-143-3p) than in the NC group (Lv-miR-NC) (Fig. 3d−f). Additionally, a Matrigel plug assay was performed to assess the effects of miR-143-3p on angiogenesis by subcutaneously injecting the mixture of Matrigel and stably expressing miR-143-3p NOZ cells into BALB/c nude mice; immunohistochemistry (IHC) using the CD31 antibody was performed to evaluate angiogenesis. The result showed that fewer vessels were formed in the miR-143-3p overexpression group (Lv-miR-143-3p) than in the NC group (Lv-miR-NC) (Fig. 3a). Furthermore, immunostaining of the CD31 protein in the Matrigel plugs of the Lv-miR-143-3p group was remarkably weaker than in the Lv-miR-NC group (Fig. 3b). The vessel densities were lower in the Lv-miR-143-3p plugs than in the Lv-miR-NC plugs (Fig. 3c).
a Matrigel containing 20 U of heparin and NOZ cells transfected with Lv-miR-NC or Lv-miR-143-3p was subcutaneously implanted in 4−6-week-old male BALB/c athymic nude mice. After 7 days, the Matrigel plugs were removed and photographed. n = 5 per group. b H&E and CD31 staining of the Matrigel plug. Scale bar, 100 mm. c Quantification of the microvessel density (mm−2; n = 5; *P < 0.05; Student’s t test). d Representative examples of tumours formed in nude mice implanted with the indicated cells. e, f The tumour growth curves are summarized in the line chart. A statistical plot of the average tumour weights in the subcutaneous xenograft model (**P < 0.01, ***P < 0.001, n = 5)
miR-143-3p inhibits PLGF-induced angiogenesis
To determine the role of miR-143-3p in angiogenesis, an angiogenesis antibody array was performed to assess differences in expression of angiogenesis-related cytokines between the conditional medium of NOZ cells transfected with either the mimic NC or miR-143-3p mimics. As shown in Fig. 4a, b, the levels of nine cytokines (CXCL5, uPAR, VEGFR2, VEGFR3, VEGFA, MMP-9, ANGPT1, TGF-β and PLGF) were lower in the NOZ miR-143-3p group compared with the NOZ mimic-NC group. Among them, PLGF was dramatically downregulated by approximately 71%. We measured the PLGF levels in the cell supernatants after transfection with the miR-143-3p mimics or mimic NC by enzyme-linked immunosorbent assay (ELISA). Consistent with the angiogenesis antibody array data, the PLGF level was significantly downregulated in the miR-143-3p mimics group compared with the mimic NC group in both NOZ and GBC-SD cells (Fig. 4c). The PLGF mRNA and protein expression levels were determined by qRT- PCR and western blot, and the results indicated that miR-143-3p inhibited the expression of PLGF at the transcriptional level (Fig. 4d, e), while silencing of miR-143-3p enhanced the expression of PLGF (Fig. 4d, e). Because miR-143-3p inhibits tube formation and invasion, we assessed whether inhibition of angiogenesis occurred via downregulation of PLGF. Hence, the recombinant PLGF protein was used to increase PLGF expression. The results showed that the addition of recombinant PLGF protein significantly increased HMVEC tube formation and invasion, whereas no significant differences were observed between the mimic NC+IgG and miR-143-3p+PLGF groups (Fig. 4f−i). No binding sites were found in the 3′UTR of PLGF, indicating that PLGF was not a direct target of miR-143-3p.
a Human angiogenesis array analysis of the conditional medium from NOZ-mimic-NC and NOZ-miR-143-3p mimic cells. b A summary of the relative expression levels of the angiogenesis cytokines is provided in the bar graph. c PLGF in the supernatants of the NOZ and GBC-SD cells that were transfected with mimic NC or the miR-143-3p mimics were quantified by ELISA (n = 3; *P < 0.05, **P < 0.01; Student’s t test). d, e PLGF expression in the mimic (inhibitor) NC and miR-143-3p mimics (inhibitors)-transfected GBC cells was analysed by western blot and qRT-PCR analysis. GAPDH was used as the loading control. f, g Endothelial tube formation estimation after incubation of HMVECs with conditioned medium from mimic NC or miR-143-3p cancer cells with or without PLGF. The number of branches was quantified (P < 0.001; Student’s t test). h, i Invasion of HMVECs through the Matrigel chambers after incubation with conditioned medium from mimic NC or miR-143-3p cancer cells with or without PLGF. Scale bar, 100 µm. The number of invading cells was determined and is depicted in the bar graph (***P < 0.001; Student’s t test)
ITGA6 is a direct target gene of miR-143-3p
To explore the molecular mechanisms by which miR-143-3p regulates GBC cell growth and angiogenesis, three miRNA target-prediction programmes (TargetScan, PicTar and miRDB) were used to predict the target sites of miR-143-3p (Fig. 5a). ITGA6 was among the predicted targets. ITGA6 is a major component of the ECM signalling pathway, which reportedly promotes tumour angiogenesis20. The 1990−1997 region of the ITGA6 3′-UTR contains a conserved miR-143-3p binding site (Fig. 5b). Thus, ITGA6 was selected as a candidate target for further analyses. qRT- PCR and western blot analyses confirmed that miR-143-3p suppressed the endogenous expression of ITGA6 at the mRNA and protein levels (Fig. 5c, d). Additionally, immunofluorescence assays showed that the expression levels of ITGA6 in the miR-143-3p mimic-transfected cells were weaker than in the miR-NC-transfected cells. Inhibition of miR-143-3p reversed this phenotype (Fig. 5f). Additionally, the ITGA6 assessment of the xenografted tumours showed comparable results by IHC (Supplementary Figure S2a). To further confirm that miR-143-3p interacted with the 3′-UTR regions of the ITGA6 mRNA at the predicted seed sequence binding sites, we constructed a dual-luciferase reporter plasmid containing a fragment of the ITGA6 3′-UTR across the conserved miR-143-3p binding sites (Fig. 5b). The dual-luciferase activity assays showed that expression of the ITGA6 reporter was significantly reduced by the miR-143-3p mimics and increased by the miR-143-3p inhibitors. Conversely, expression of the ITGA6 reporter containing the mutated sequence of the same fragment was not affected by the miR-143-3p mimics or inhibitors (Fig. 5e). These results indicate that ITGA6 is a direct downstream target of miR-143-3p.
a Potential miR-143-3p targets predicted by the three miRNA target-prediction programmes (TargetScan, PicTar and miRDB). b The wild-type or mutant ITGA6 3′UTRs to determine the miR-143-3p binding site. c, d Overexpression of miR-143-3p attenuates ITGA6 protein and mRNA expression in NOZ and GBC-SD cells compared with the miRNA-NC group. Silencing of miR-143-3p promotes ITGA6 protein and mRNA expression in NOZ and GBC-SD cells compared with the miRNA-NC group. (*P < 0.05, **P < 0.01; Student’s t test). e The relative luciferase activity of the wild-type or mutant ITGA6 3′UTR in 293T cells after transfections with the miR-143-3p mimic or inhibitor and corresponding control (*P < 0.05, NS no significant). f Expression of ITGA6 in GBC cells infected with the miR-143-3p mimic or inhibitor and corresponding control was examined by immunofluorescence. The red signals represent ITGA6 staining. The nuclei were counterstained with DAPI
ITGA6 expression is upregulated and negatively associated with miR-143-3p expression in GBC
To further evaluate the correlation between ITGA6 and miR-143-3p in GBC tissues, we examined the levels of ITGA6 mRNA in the same 49 pairs of GBC tissues and their corresponding NATs. The qRT-PCR results showed that ITGA6 expression was significantly higher in the GBC tumour tissues than in the corresponding NATs (Fig. 6a and Supplementary Figure S2b). Furthermore, the correlation between miR-143-3p and ITGA6 in the GBC tissues was evaluated using the Spearman correlation analysis. The Spearman correlation analysis clearly showed a negative correlation between ITGA6 and miR-143-3p expression in the GBC tissues (Fig. 6b). To further evaluate the negative correlation between ITGA6 and miR-143-3p, immunohistochemical staining of the tissues with high and low levels of miR-143-3p was performed, and the results showed that the tissues with high miR-143-3p expression had weaker ITGA6 staining than the tissues with low miR-143-3p expression (Fig. 6c). Additionally, the average staining score for ITGA6 expression was higher in the miR-143-3p-low group than in the miR-143-3p-high group (**P < 0.01; Fig. 6d). Moreover, the Kaplan−Meier analysis revealed that higher ITGA6 expression had a short overall survival trend (P = 0.0176; Fig. 6e).
a The ITGA6 mRNA levels in 49 pairs of GBC tissues and their corresponding NATs. b The correlation between the expression levels of miR-143-3p and ITGA6 was determined using a linear regression analysis and a paired t test with the same samples used (P < 0.001, r = −0.2177, n = 49; Pearson’s correlation). c Representative IHC micrographs showing ITGA6 protein expression in GBC tissues with high or low miR-143-3p expression. Scale bar, 100 µm. d Scatterplots of the average staining scores of ITGA6 expression in the miR-143-3p-high or miR-143-3p-low tissues (**P < 0.01, n = 25 for miR-143-3p-low group; n = 24 for miR-143-3p high group). (e) Kaplan−Meier overall survival curve of GBC patients based on ITGA6 expression (P < 0.05, n = 49)
ITGA6 overexpression rescues the effects of miR-143-3p on GBC cell proliferation and angiogenesis
Rescue experiments were performed to further assess whether the effects of miR-143-3p on GBC cell angiogenesis and proliferation were indeed mediated by ITGA6. NOZ and GBC-SD cells were co-transfected with miR-NC or the miR-143-3p mimics together with an empty vector or ITGA6 vector. The results showed that overexpression of ITGA6 partially reversed the phenotype caused by overexpression of miR-143-3p (Fig. 7a−e). Furthermore, to determine whether ITGA6 regulated the expression of PLGF, western blot and ELISA were performed to assess PLGF levels in the co-transfected cells. The results showed that the levels of PLGF increased in the GBC cells transfected with miR-143-3p and ITGA6 compared with the GBC cells co-transfected with miR-143-3p and empty vector (Fig. 7g, f). Thus, ITGA6 overexpression can reverse downregulation of PLGF by miR-143-3p.
a, c Endothelial tube formation was estimated following incubation of HMVECs with conditioned medium from GBC cells transfected with mimic NC or miR-143-3p mimic and empty vector or ITGA6. The number of branches was quantified (*P < 0.05, **P < 0.01, ***P < 0.001; Student’s t test). Scale bar, 100 µm. b, d Invasion of HMVECs through the Matrigel chambers after incubation with conditioned medium from GBC cells co-transfected with mimic NC or miR-143-3p mimic and empty vector or ITGA6 for 48−72 h. Scale bar, 100 µm. The number of invading cells was determined and is depicted in the bar graph (**P < 0.01, ***P < 0.01; Student’s t test). e The growth rates of the GBC cells that were co-transfected with mimic NC or miR-143-3p mimics and empty vectors or ITGA6 were determined with CCK-8 proliferation assays (**P < 0.01, ***P < 0.001). f PLGF in the supernatants from NOZ and GBC-SD cells that were co-transfected with mimic NC or miR-143-3p mimic and empty vector or ITGA6 was quantified by ELISA (n = 3; *P < 0.05, ***P < 0.001; Student’s ttest). g Western blot for the ITGA6 and PLGF proteins in GBC cells that were co-transfected with mimic NC or miR-143-3p mimic and empty vector or ITGA6
miR-143-3p downregulates PLGF expression through the ITGA6/PI3K/AKT /STAT3 pathways
ITGA6 activates multiple signal transduction cascades, including PI3K/AKT and MAPK/ERK20. To investigate the molecular mechanism underlying the angiogenesis controlled by miR-143-3p/ITGA6, we examined the expression levels of relevant proteins in the MAPK/ERK and PI3K/AKT pathways. The expression levels of p-ERK1/2, p-MEK1/2, PIK3CA, p-AKT and p-STAT3 were decreased when miR-143-3p was overexpressed (Fig. 8a left). Conversely, the expression levels of p-ERK1/2, p-MEK1/2, PIK3CA, p-AKT and p-STAT3 were increased when miR-143-3p was silenced (Fig. 8a right). These changes were partially recovered by overexpression of ITGA6, indicating that miR-143-3p/ITGA6 functions as a key mediator of angiogenesis via the MAPK/ERKand PI3K/AKT /STAT3 signalling pathways. Moreover, STAT3 can bind to the promoter of PLGF to enhance its expression21. We decreased STAT3 expression by transfecting small interfering RNA (siRNA) against STAT3. After siRNA transfection, the expression of PLGF was significantly downregulated, which was consistent with the conclusion that STAT3 enhanced PLGF expression (Supplementary Figure S3a-b). Therefore, these findings suggest that miR-143-3p downregulates the expression of PLGF through the ITGA6/PI3K/AKT pathways (Fig. 8c).
a Western blot analysis of relevant proteins in the MAPK/ERK and PI3K/AKT pathways in GBC cells that were transfected with mimic (inhibitor) NC or miR-143-3p mimics (inhibitors). b Western blot analysis of relevant proteins in the MAPK/ERK and PI3K/AKT pathways in GBC cells that were co-transfected with mimic NC or miR-143-3p mimic and empty vector or ITGA6. c Working model of the miR-143-3p regulatory axis in GBC
Discussion
Accumulative evidence indicates that miRNAs serve as oncogenes or tumour suppressors that act as pivotal players in tumourigenesis and tumour progression. Additionally, deregulation of miRNAs has been observed in various cancers, including gastric cancer, liver cancer, pancreatic cancer and colorectal cancer22,23,24,25. Numerous studies have examined the expression profiles of miRNAs to explore the mechanism of miRNA actions in tumourigenesis and tumour progression. Moreover, new studies have established the potential usefulness of miRNAs as therapeutic molecules against cancer25. According to our miRNA profiling results1, we found that miR-143-3p was significantly downregulated. We further confirmed this observation in 49 pairs of GBC tissues. Our results further indicated that miR-143-3p inactivated the MAPK/ERK and PI3K/AKT/STAT3 pathways in GBC cells by directly targeting ITGA6, resulting in a decrease in PLGF and the development of GBC angiogenesis and tumour growth. Therefore, our results reveal a leading mechanism by which miR-143-3p suppresses angiogenesis in GBC.
miR-143-3p acts as a tumour suppressor in various cancers, including lung cancer, colorectal cancer, prostate cancer and hepatocellular carcinoma26,27,28,29. Low expression of miR-143-3p is associated with a poor clinical outcome. Importantly, increasing evidence has shown the potential usefulness of miRNAs as therapeutic molecules against cancer. Circulating levels of miRNAs are also quite stable and reproducible in the body fluids of many cancer patients. Given the critical roles of miRNAs in tumour development and in physiological regulation overall30, further characterizations and screenings of therapeutic targets for GBC hold great promise. However, the precise molecular mechanisms by which miR-143-3p influences various physiological processes of GBC remain unknown. To date, biological functions for miR-143-3p have been reported mainly in areas of proliferation and metastasis, while the impact of miR-143-3p on angiogenesis is not well understood. Therefore, we focused on angiogenesis and performed in vivo and in vitro angiogenesis assays to explore the effects of miR-143-3p on angiogenesis. Through gain- and loss-of-function miR-143-3p assays in GBC cells, we uncovered that miR-143-3p could efficiently weaken in vitro invasion and capillary tube formation of HMVECs and suppress tumour growth and angiogenesis in nude mice. Our results indicate that miR-143-3p is a novel antiangiogenesis miRNA. Angiogenesis has a pivotal function in tumour growth and progression and is responsible for the rapid recurrences of tumours and poor prognoses for patients. Numerous factors are involved in angiogenesis. Among them, VEGF plays an important role in various cancers. To determine the target of miR-143-3p that was relevant to inhibition of angiogenesis, an angiogenesis antibody array was performed, and the results indicated that several pro-angiogenesis cytokines were downregulated by miR-143-3p, including CXCL5, uPAR, VEGFR2, VEGFR3, VEGFA, MMP-9, ANGPT1, TGF-β and PLGF. Among them, PLGF showed a sharp decrease. The ELISA and western blot analyses also confirmed these results. PLGF is a member of the VEGF family. Multiple reports have described the relationship between PLGF, angiogenesis and metastasis in cancer. For example, PLGF promotes metastasis of non-small-cell lung cancer through MMP931. To determine whether miR-143-3p directly bound to PLGF, we compared the miR-143-3p sequence with the PLGF 3′UTR and found no binding site. We presumed that miR-143-3p decreased the expression of PLGF by interacting with an upstream regulator of PLGF. Therefore, a bioinformatics prediction system (TargetScan, PicTar and miRDB) was used to predict the target of miR-143-3p. According to the results, ITGA6 was a direct target of miR-143-3p and was associated with angiogenesis. Therefore, we hypothesized that miR-143-3p decreased the expression of PLGF by interacting with ITGA6. We confirmed this hypothesis with dual-luciferase assays and rescue assays, with follow-up qRT-PCR and western blot assays to further validate the results.
ITGA6 (integrin α6), also known as CD49f, is a member of the integrin family that plays a vital role in the interactions between many cell types and is involved in several biological processes, including cell survival, proliferation and gene transcription32. ITGA6 associates with the integrin β1 chain and integrin β4 to form the α6β1 or α6β4 complex. α6β1 and α6β4 are important receptors for laminin and are essential for the ECM pathways20. Interest in defining the contribution of integrins to transcription has been high. Furthermore, the ability of integrins to regulate translation provides a mechanistic basis for altering cell functions by increasing the expression levels of specific proteins30. Recent studies have suggested that ITGA6 regulates eIF-4E activity and VEGF translation33. Moreover, ITGA6 is associated with invasion, metastasis and a poor prognosis in human GBC32,34,35. In this study, we have shown that ITGA6 indirectly downregulates PLGF via the PI3K/AKT/STAT3 pathway. Additionally, activation of AKT and STAT3 plays an essential role in tumour development and progression4. Moreover, we performed rescue, western blot and ELISA analyses to confirm that miR-143-3p decreased the expression levels of PLGF by interacting with ITGA6. ITGA6 overexpression could markedly reverse the inhibitory effects of miR-143-3p on GBC cell proliferation and angiogenesis and on PLGF expression. Thus, the antiangiogenic functions of miR-143-3p are indirectly executed by inhibiting its indirect target, PLGF.
In summary, our study demonstrated the antiangiogenic effects of miR-143-3p on GBC cells. Furthermore, our study characterized a novel mechanism that underlies the effects of miR-143-3p on GBC cells: the miR-143-3p/ITGA6/PLGF axis. Collectively, our results indicate that miR-143-3p and its target ITGA6 may be effective prognostic indicators. Additionally, more studies of miR-143-3p and ITGA6 should be conducted so that new approaches for molecular therapeutics that specifically target miR-143-3 and ITGA6 in GBC patients can be developed.
Materials and methods
Tissue samples
Human GBC samples and the corresponding normal gallbladder tissues were obtained from the Department of General Surgery, Xinhua Hospital (Shanghai, China). Additionally, this study was approved by the Research Ethics committee. Written informed consent was obtained from all participants. Tissue samples were collected during surgery and immediately frozen in liquid nitrogen.
Cell culture and reagents
The GBC-SD and SGC996 human GBC cell lines that were used in this study were purchased from the cell bank of the type culture collection of the Chinese Academy of Sciences (Shanghai, China). NOZ, OCUG-1 and EHGB-1 cells were obtained from the Health Science Research Bank (Osaka, Japan). HMVECs were provided by Dr. Rong Shao36. NOZ, GBC-SD, EHGB-1 and OCUG-1 cells were maintained in high-glucose DMEM (Gibco, USA), and HMVECs were maintained in ECM (ScienCell, USA) supplemented with 10% FBS (Gibco, USA), penicillin G (100 U/ml) and streptomycin (100 g/ml). The SGC996 cells were cultured in Roswell Park Memorial Institute (Gibco, USA) 1640 medium supplemented with 10% FBS, penicillin G (100 U/ml) and streptomycin (100 g/ml). Cells were maintained as monolayer cultures at 37 °C in humidified air with 5% CO2 and 95% air. Before the experiments, cell lines were validated by microscopy, growth curve analysis and mycoplasma detection according to the cell line verification test recommendations.
RNA extraction and quantitative real-time PCR
Total RNA was extracted from the tissue samples or cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). Quantitative real-time PCR was performed using SYBR® Green (Takara, Japan) according to the manufacturer’s instructions. The qRT-PCR results were analysed and examined as the relative miRNA or mRNA levels based on cycle threshold (CT) values, which were converted to fold changes. The primer sequences used are listed in Supplementary Table S1.
Cell transfection
Hsa-miRNA mimics, hsa-miRNA inhibitors and their cognate control RNAs were purchased from Riobio (Guangzhou, China). Lentivirus-miR-143-3p and lentivirus-miR-NC were purchased from Genomeditech (Shanghai, China). Additionally, an ITGA6 overexpression plasmid was used for the rescue experiments. The miRNA mimics, miRNA inhibitors and plasmids were transfected into NOZ and GBC-SD cells using Lipofectamine 2000 transfection reagent (Invitrogen, USA). Total RNA and protein were collected 48 h after transfection.
Generation of stable cell lines with overexpression of miR-143-3p
For the construction of cell lines stably expressing miR-143-3p, miR-143-3p and negative control sequence were synthesized and inserted into PGMLV-hU6-MCS-CMV-ZsGreen1-PGK-puromycin-WPRE lentiviral vector. Recombinant lentiviruses expressing miR-143-3p or negative control (Lv-miR-143-3p and Lv-miR-NC, respectively) were produced by Genomeditech (Shanghai, China). NOZ cells were infected with concentrated virus, and the culture medium was replaced after 24 h incubation. Then, cells were treated with 1 μg/ml puromycin for 2 weeks for the selection of stable cell lines. The expression of miR-143-3p in the stable cell lines and xenograft tumour was validated by qRT–PCR analysis.
Human angiogenesis antibody array
The human angiogenesis antibody array kit was purchased from RayBiotech (AAH-ANG-1000-4, Norcross, GA). A total of 1×105 NOZ cells transfected with mimic NC or miR-143-3p mimics were seeded onto a six-well plate with 1 ml of serum-free medium. Forty-eight hours later, the conditioned medium was collected and used for a human angiogenesis antibody array analysis. The procedure was performed as per the manufacturer’s instructions. Briefly, 2 ml of blocking buffer was pipetted into the angiogenesis antibody membrane and incubated for 30 min at room temperature (RT). After the incubation, the blocking buffer was aspirated. The antibody membrane was washed twice with the array wash buffer. Afterwards, 1 ml of conditioned medium was added into the well, and the antibody membranes were incubated overnight at 4 °C. Two millilitres of the HRP-Streptavidin solution was added onto the membrane and incubated for 2 h at RT. A chemiluminescent substrate ECL kit was used to obtain detailed pictures of the array.
Enzyme-linked immunosorbent assay
Enzyme-linked immunosorbent assay was used to measure the levels of PLGF in the cancer cell supernatants. The human PLGF ELISA kit (Boster, Wuhan, China) was used according to the manufacturer’s instructions.
In vitro tube formation and invasion assay
Ten microliters of ice-cold Matrigel (BD MatrigelTM, USA) was added into culture plates (µ-Slide Angiogenesis, ibidi, USA). Cells (2×104/well) in 50 μl of conditioned medium were then seeded onto these plates. After incubation for 4–6 h at 37 °C, Calcein AM (Sigma, USA) was added to the plates. Tube formation was examined using photographs obtained from fluorescence microscopy examinations (Leica, Germany). The number of branch points was determined and analysed in five random fields per replicate. For the invasion assays, HMVECs (2×104/well) in 200 μl of FBS-free DMEM were added into the upper chambers, while the lower chambers were filled with 600 μl of conditioned medium. After 48−72 h of incubation, the filters were removed and fixed with 4% paraformaldehyde for 30 min. The cells located on the bottom sides of the filters were stained with crystal violet for 20 min. The stained cells that had invaded were counted in five randomly chosen fields (×100 magnification) per well.
Subcutaneous xenograft models
The subcutaneous xenograft model was established using 4- to 6-week-old male BALB/c athymic nude mice. The mice were purchased from the Shanghai Laboratory Animal Center of the Chinese Academy of Sciences (Shanghai, China). The mice were housed under specific pathogen-free conditions and fed a regular autoclaved chow diet with water ad libitum. A total of 1×106 NOZ cells stably expression miR-143-3p or negative control (Lv-miR-143-3p or Lv-miR-NC) were inoculated subcutaneously into the ventral areas of the mice (n = 5 per group). The sizes of the tumours were measured every 2 days after inoculation. Additionally, at the endpoint (approximately 3 weeks), the weights of the tumours were measured. The sizes were evaluated using the following formula: tumour volume = (tumour length × (tumour width)2) × 0.52. After that, the tumours were fixed and used for in situ hybridization (ISH) and IHC assays. All animal experiments were approved by the Institutional Animal Care and Use Committee of Xinhua Hospital (2013-0106) and conducted humanely.
In vivo Matrigel plug assay
NOZ cells were infected with a negative control lentivirus or a mir-143-3p lentivirus (Lv-miR-NC or Lv-miR-143-3p) and were selected with 0.5 µg/ml of puromycin for 10 days. A total of 0.5 ml of Matrigel (BD Biosciences, USA) containing 20 U of heparin and 1×106 infected NOZ cells was injected subcutaneously into the ventral areas of BALB/c athymic nude mice (n = 5 per group). After 7 days, mice were killed, and the plugs were removed. The plugs were photographed, fixed and stained for H&E and CD31 analyses.
Luciferase reporter assay
To investigate the binding site of miR-143-3p and its candidate target gene ITGA6, dual-luciferase reporter assays were performed using 293T cells with the pmirGLO System (Promega, USA) following the manufacturer’s protocol. Cells were co-transfected with 50 nM miR-143-3p mimic, the inhibitor or the cognate controls and 0.5 μg of pmirGLO-ITGA6-3′UTR-WT/pmirGLO-ITGA6-3′UTR-MUT vector. After 48 h, cells were collected, and the luciferase activity was analysed according to the manufacturer’s protocol (Promega, USA).
Immunofluorescence
After transfections with the mimics or inhibitors, GBC cells were seeded onto 24-well plates. Two days later, the cells were washed with PBS and fixed in 4% for 30 min at RT. Afterwards, cells were incubated with blocking buffer (5% BSA, 0.1% Triton X-100) alone for 1 h and with the primary antibodies overnight at 4 °C. The primary antibody was removed, and cells were incubated with the secondary fluorescent antibody (594) for 1 h at RT. After several PBS washes, cells were stained with DAPI and photographed under a fluorescence microscope (Leica, Germany).
Immunohistochemistry
Immunohistochemistry was performed using the anti-ITGA6 (Sigma, 1:200), anti-Ki67 (Proteintech, 1:400), anti-PLGF (Abcam, 1:200) and anti-mouse CD31 (Abcam, 1:400) antibodies. Frozen or paraffin-embedded sections were used for immunohistochemical analysis. For immunohistochemical staining, tissue sections were first incubated with 0.1% trypsin at RT for 10 min followed by incubation with 0.1 μg/ml trypsin inhibitor (Sigma) for 5 min. The sections were then rinsed three times in PBS before blocking with 10% normal goat serum (15 min at RT). The sections were rinsed in PBS and incubated with a primary antibody overnight at 4 °C. Afterwards, the tissue sections were rinsed three times in PBS and incubated with the secondary antibody for 60 min at RT. The sections were then immersed in DAB for 5−10 min and counterstained with 10% Mayer’s haematoxylin. ITGA6 expression in tissues was evaluated according to the methods described by R Shao et al 37.
In situ hybridization
The triple digoxigenin-labelled antisense locked nucleic acid (LNA)-modified probes for miR-143-3p were synthesized by Boster Biotech (Wuhan, China). ISH was conducted according to the manufacturer’s instruction of microRNA ISH Optimization Kits (Boster, Wuhan, China).
Statistical analysis
All experiments were repeated three times unless otherwise noted. The data are presented as the mean ± SD. Student’s t test was used for single comparisons, and one-way ANOVA was used for multiple comparisons. Kaplan−Meier curves and log-rank tests were performed to compare the survival times among the groups. All data were analysed with GraphPad Prism 5 and SPSS v17.0 software. P < 0.05 was considered statistically significant.
References
Shu, Y. et al. MicroRNA-29c-5p suppresses gallbladder carcinoma progression by directly targeting CPEB4 and inhibiting the MAPK pathway. Cell Death Differ. 24, 445–457 (2017).
Gourgiotis, S. et al. Gallbladder cancer. Am. J. Surg. 196, 252–264 (2008).
Shu, Y. et al. SPOCK1 as a potential cancer prognostic marker promotes the proliferation and metastasis of gallbladder cancer cells by activating the PI3K/AKT pathway. Mol. Cancer 14, 12 (2015).
Zhang, Y. et al. A novel PI3K/AKT signaling axis mediates Nectin-4-induced gallbladder cancer cell proliferation, metastasis and tumor growth. Cancer Lett. 375, 179–189 (2016).
Hundal, R. & Shaffer, E. Gallbladder cancer: epidemiology and outcome. Clin. Epidemiol. 6, 99–109 (2014).
Li, M. et al. Whole-exome and targeted gene sequencing of gallbladder carcinoma identifies recurrent mutations in the ErbB pathway. Nat. Genet. 46, 872–876 (2014).
Li, Z. et al. LASP-1 induces proliferation, metastasis and cell cycle arrest at the G2/M phase in gallbladder cancer by down-regulating S100P via the PI3K/AKT pathway. Cancer Lett. 372, 239–250 (2016).
Wu, S. et al. A miR-192-EGR1-HOXB9 regulatory network controls the angiogenic switch in cancer. Nat. Commun. 7, 11169 (2016).
De Palma, M., Biziato, D. & Petrova, T. Microenvironmental regulation of tumour angiogenesis. Nat. Rev. Cancer 17, 457–474 (2017).
Hicklin, D. & Ellis, L. Role of the vascular endothelial growth factor pathway in tumor growth and angiogenesis. J. Clin. Oncol. 23, 1011 (2005).
De Falco, S. The discovery of placenta growth factor and its biological activity. Exp. Mol. Med. 44, 1–9 (2012).
Lee, J. et al. Safety and clinical activity of the programmed death-ligand 1 inhibitor durvalumab in combination with poly (ADP-ribose) polymerase inhibitor olaparib or vascular endothelial growth factor receptor 1-3 inhibitor cediranib in women’s cancers: a dose-escalation, phase I study. J. Clin. Oncol. 35, 2193–2202 (2017).
Faivre, S. et al. Sunitinib in pancreatic neuroendocrine tumors: updated progression-free survival and final overall survival from a phase III randomized study. Ann. Oncol. 28, 339–343 (2017).
Hatem, R. et al. Vandetanib as a potential new treatment for estrogen receptor-negative breast cancers. Int. J. Cancer 138, 2510–2521 (2016).
Liang, Y. et al. The EGFR/miR-338-3p/EYA2 axis controls breast tumor growth and lung metastasis. Cell Death Dis. 8, e2928 (2017).
Liu, K. et al. SOX2 regulates multiple malignant processes of breast cancer development through the SOX2/miR-181a-5p, miR-30e-5p/TUSC3 axis. Mol. Cancer 16, 62 (2017).
Liu, Y. et al. miR-19a promotes colorectal cancer proliferation and migration by targeting TIA1. Mol. Cancer 16, 53 (2017).
Li, G. & Pu, Y. MicroRNA signatures in total peripheral blood of gallbladder cancer patients. Tumour Biol. 36, 6985–6990 (2015).
Letelier, P. et al. miR-1 and miR-145 act as tumor suppressor microRNAs in gallbladder cancer. Int. J. Clin. Exp. Pathol. 7, 1849–1867 (2014).
Stewart, R. & O’Connor, K. Clinical significance of the integrin α6β4 in human malignancies. Lab Invest. 95, 976–986 (2015).
Tu, H. et al. Enhancement of placenta growth factor expression by oncostatin M in human rheumatoid arthritis synovial fibroblasts. J. Cell Physiol. 228, 983–990 (2013).
Han, T. et al. MicroRNA-29c mediates initiation of gastric carcinogenesis by directly targeting ITGB1. Gut 64, 203–214 (2015).
Chang, R. et al. MicroRNA-331-3p promotes proliferation and metastasis of hepatocellular carcinoma by targeting PH domain and leucine-rich repeat protein phosphatase. Hepatology 60, 1251–1263 (2014).
Imamura, T. et al. Depleted tumor suppressor miR-107 in plasma relates to tumor progression and is a novel therapeutic target in pancreatic cancer. Sci. Rep. 7, 5708 (2017).
Xiao, J. et al. Therapeutic inhibition of miR-4260 suppresses colorectal cancer via targeting MCC and SMAD4. Theranostics 7, 1901–1913 (2017).
Liu, X., Gong, J. & Xu, B. miR-143 down-regulates TLR2 expression in hepatoma cells and inhibits hepatoma cell proliferation and invasion. Int. J. Clin. Exp. Pathol. 8, 12738–12747 (2015).
Su, J. et al. MiR-143 and MiR-145 regulate IGF1R to suppress cell proliferation in colorectal cancer. PLoS ONE 9, e114420 (2014).
Wei, J. et al. miR-143 inhibits cell proliferation by targeting autophagy-related 2B in non-small cell lung cancer H1299 cells. Mol. Med. Rep. 11, 571–576 (2015).
Wu, D. et al. MicroRNA-143 inhibits cell migration and invasion by targeting matrix metalloproteinase 13 in prostate cancer. Mol. Med. Rep. 8, 626–630 (2013).
Li, L. et al. Serum miRNAs as predictive and preventive biomarker for pre-clinical hepatocellular carcinoma. Cancer Lett. 373, 234–240 (2016).
Zhang, W. et al. Placental growth factor promotes metastases of non-small cell lung cancer through MMP9. Cell Physiol. Biochem. 37, 1210–1218 (2015).
Lowell, C. & Mayadas, T. Overview: studying integrins in vivo. Methods Mol. Biol. 757, 369–397 (2012).
Chung, J. et al. Integrin (alpha 6 beta 4) regulation of eIF-4E activity and VEGF translation: a survival mechanism for carcinoma cells. J. Cell Biol. 158, 165–174 (2002).
Zhang, D. et al. Overexpression of Thy1 and ITGA6 is associated with invasion, metastasis and poor prognosis in human gallbladder carcinoma. Oncol. Lett. 12, 5136–5144 (2016).
Pabla, R. et al. Integrin-dependent control of translation: engagement of integrin alphaIIbbeta3 regulates synthesis of proteins in activated human platelets. J. Cell Biol. 144, 175–184 (1999).
Shao, R. & Guo, X. Human microvascular endothelial cells immortalized with human telomerase catalytic protein: a model for the study of in vitro angiogenesis. Biochem. Biophys. Res. Commun. 321, 788–794 (2004).
Shao, R. et al. YKL-40, a secreted glycoprotein, promotes tumor angiogenesis. Oncogene 28, 4456–4468 (2009).
Acknowledgements
This study was supported by the National Natural Science Foundation of China (No. 31620103910, 91440203, 81402403, 81502433, 31601021 and 31501127), the National High Technology Research and Development Program (863 Program) (No. 2012AA022606), the Shanghai Science and Technology Commission Key Basic Research Program (No. 16JC1400200), the Key Program of Shanghai Science and Technology Commission (No. 16411952501), the Program for Changjiang Scholars, the Multiple Central Clinical Research Program of Shanghai Jiao Tong University School of Medicine (No. DLY201507), the Precision Medicine Research Program of Shanghai Jiao Tong University School of Medicine (No. 15ZH4003), the Leading Talent Program of Shanghai and Specialized Research Foundation for the PhD Program of Higher Education-Priority Development Fields (No. 20130073130014), the Interdisciplinary Program of Shanghai Jiao Tong University (No. 14JCRY05), the Shanghai Rising Stars Program (No. 15QA1403100).
Authors’ contributions
Y.-B.L., Y.-P.J. and Y.-P.H. were responsible for the experimental design. Y.-P.J., Y.-P.H., X.-S.W., Y.-S.W. carried out most experiments in this work. Y.-P.J., Y.-P.H., Y.-Y.Y., Y.-C.L. participated in performing the experiment and in the manuscript mapping and submission. L.J., Y.-J.Z., Y.-J.H., F.-T.L., H.-F.L. participated in the discussion and data interpretation. Y.-B.L. was responsible for the funding application and the supervision and management of the project. All authors have read and approved the final manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Jin, YP., Hu, YP., Wu, XS. et al. miR-143-3p targeting of ITGA6 suppresses tumour growth and angiogenesis by downregulating PLGF expression via the PI3K/AKT pathway in gallbladder carcinoma. Cell Death Dis 9, 182 (2018). https://doi.org/10.1038/s41419-017-0258-2
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41419-017-0258-2
This article is cited by
-
Topological reorganization and functional alteration of distinct genomic components in gallbladder cancer
Frontiers of Medicine (2024)
-
tRF-3013b inhibits gallbladder cancer proliferation by targeting TPRG1L
Cellular & Molecular Biology Letters (2022)
-
Identification of invasion-metastasis associated MiRNAs in gallbladder cancer by bioinformatics and experimental validation
Journal of Translational Medicine (2022)
-
Non-coding RNAs as potential biomarkers of gallbladder cancer
Clinical and Translational Oncology (2022)
-
Association study between immune-related miRNAs and mixed connective tissue disease
Arthritis Research & Therapy (2021)