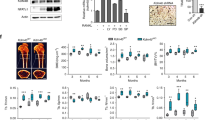
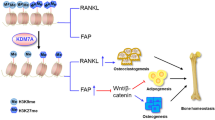
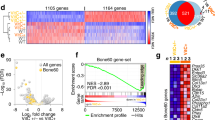
Abstract
Absent, small, or homeotic1-like (ASH1L) is a histone lysine methyltransferase that generally functions as a transcriptional activator in controlling cell fate. So far, its physiological relevance in bone homeostasis and osteoclast differentiation remains elusive. Here, by conditional deleting Ash1l in osteoclast progenitors of mice, we found ASH1L deficiency resulted in osteoporosis and potentiation of osteoclastogenesis in vivo and in vitro. Mechanistically, ASH1L binds the promoter of the Src homology 3 and cysteine-rich domain 2 (Stac2) and increases the gene’s transcription via histone 3 lysine 4 (H3K4) trimethylation modification, thus augmenting the STAC2’s protection against receptor activator of nuclear factor kB ligand (RANKL)-initiated inflammation during osteoclast formation. Collectively, we demonstrate the first piece of evidence to prove ASH1L as a critical checkpoint during osteoclastogenesis. The work sheds new light on our understanding about the biological function of ASH1L in bone homeostasis, therefore providing a valuable therapeutic target for the treatment of osteoporosis or inflammatory bone diseases.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
RNA-seq data and CUT&Tag-seq data have been deposited at the NCBI Sequence Read Archive at https://www.ncbi.nlm.nih.gov/sra with accession number PRJNA947264 and PRJNA1065086. Other data in this study are available upon request from the corresponding author.
References
Park JH, Lee NK, Lee SY. Current understanding of RANK signaling in osteoclast differentiation and maturation. Mol Cells. 2017;40:706–13.
Kurotaki D, Yoshida H, Tamura T. Epigenetic and transcriptional regulation of osteoclast differentiation. Bone.2020;138:115471.
Oton-Gonzalez L, Mazziotta C, Iaquinta MR, Mazzoni E, Nocini R, Trevisiol L, et al. Genetics and epigenetics of bone remodeling and metabolic bone diseases. Int J Mol Sci. 2022;23:1500.
Gregory GD, Vakoc CR, Rozovskaia T, Zheng X, Patel S, Nakamura T, et al. Mammalian ASH1L is a histone methyltransferase that occupies the transcribed region of active genes. Mol Cell Biol. 2007;27:8466–79.
Jones M, Chase J, Brinkmeier M, Xu J, Weinberg DN, Schira J, et al. Ash1l controls quiescence and self-renewal potential in hematopoietic stem cells. J Clin Investig. 2015;125:2007–20.
Byrd KN, Shearn A. ASH1, a Drosophila trithorax group protein, is required for methylation of lysine 4 residues on histone H3. Proc Natl Acad Sci USA. 2003;100:11535–40.
Maritz C, Khaleghi R, Yancoskie MN, Diethelm S, Brülisauer S, Ferreira NS, et al. ASH1L-MRG15 methyltransferase deposits H3K4me3 and FACT for damage verification in nucleotide excision repair. Nat Commun. 2023;14:3892.
Trissal MC, Wong TN, Yao J-C, Ramaswamy R, Kuo I, Baty J, et al. MIR142 loss-of-function mutations derepress ASH1L to increase hoxa gene expression and promote leukemogenesis. Cancer Res. 2018;78:3510–21.
Xia M, Liu J, Wu X, Liu S, Li G, Han C, et al. Histone methyltransferase Ash1l suppresses interleukin-6 production and inflammatory autoimmune diseases by inducing the ubiquitin-editing enzyme A20. Immunity.2013;39:470–81.
Xia M, Liu J, Liu S, Chen K, Lin H, Jiang M, et al. Ash1l and lnc-Smad3 coordinate Smad3 locus accessibility to modulate iTreg polarization and T cell autoimmunity. Nat Commun. 2017;8:15818.
Du WJ, Yang H, Tong F, Liu S, Zhang C, Chen Y, et al. Ash1L ameliorates psoriasis via limiting neuronal activity-dependent release of miR-let-7b. Br J Pharmacol. 2023; https://doi.org/10.1111/bph.16254.
Szulc P, Naylor K, Hoyle NR, Eastell R, Leary ET, for the National Bone Health Alliance Bone Turnover Marker Project. Use of CTX-I and PINP as bone turnover markers: National Bone Health Alliance recommendations to standardize sample handling and patient preparation to reduce pre-analytical variability. Osteoporos Int. 2017;28:2541–56.
Kodama J, Kaito T. Osteoclast multinucleation: review of current literature. Int J Mol Sci. 2020;21:5685.
Bhattarai G, Kook SH, Kim JH, Poudel SB, Lim SS, Seo YK, et al. COMP-Ang1 prevents periodontitic damages and enhances mandible bone growth in an experimental animal model. Bone. 2016;92:168–79.
Bozec A, Zaiss MM, Kagwiria R, Voll R, Rauh M, Chen Z, et al. T cell costimulation molecules CD80/86 inhibit osteoclast differentiation by inducing the IDO/tryptophan pathway. Sci Transl Med. 2014;6:235ra60.
Bradley EW, Ruan MM, Oursler MJ. Novel pro-survival functions of the Kruppel-like transcription factor Egr2 in promotion of macrophage colony-stimulating factor-mediated osteoclast survival downstream of the MEK/ERK pathway. J Biol Chem. 2008;283:8055–64.
Choi B, Kang SS, Kang SW, Min BH, Lee EJ, Song DH, et al. Secretory clusterin inhibits osteoclastogenesis by attenuating M-CSF-dependent osteoclast precursor cell proliferation. Biochem Biophys Res Commun. 2014;450:105–9.
Hu YS, Zhou H, Myers D, Quinn JM, Atkins GJ, Ly C, et al. Isolation of a human homolog of osteoclast inhibitory lectin that inhibits the formation and function of osteoclasts. J Bone Min Res. 2004;19:89–99.
Huynh H, Wei W, Wan Y. mTOR Inhibition subdues milk disorder caused by maternal VLDLR loss. Cell Rep. 2017;19:2014–25.
Jeong E, Choi HK, Park JH, Lee SY. STAC2 negatively regulates osteoclast formation by targeting the RANK signaling complex. Cell Death Differ. 2018;25:1364–74.
Kim HJ, Hong JM, Yoon KA, Kim N, Cho DW, Choi JY, et al. Early growth response 2 negatively modulates osteoclast differentiation through upregulation of Id helix-loop-helix proteins. Bone. 2012;51:643–50.
Kim JH, Kim K, Kim I, Seong S, Kim N. c-Src-dependent and -independent functions of matk in osteoclasts and osteoblasts. J Immunol. 2018;200:2455–63.
Kim WS, Kim HJ, Lee ZH, Lee Y, Kim HH. Apolipoprotein E inhibits osteoclast differentiation via regulation of c-Fos, NFATc1 and NF-κB. Exp Cell Res. 2013;319:436–46.
Li W, Wei C, Xu L, Yu B, Chen Y, Lu D, et al. Schistosome infection promotes osteoclast-mediated bone loss. PLoS Pathog. 2021;17:e1009462.
Fan R, Toubal A, Goñi S, Drareni K, Huang Z, Alzaid F, et al. Loss of the co-repressor GPS2 sensitizes macrophage activation upon metabolic stress induced by obesity and type 2 diabetes. Nat Med. 2016;22:780–91.
Soldi M, Mari T, Nicosia L, Musiani D, Sigismondo G, Cuomo A, et al. Chromatin proteomics reveals novel combinatorial histone modification signatures that mark distinct subpopulations of macrophage enhancers. Nucleic Acids Res. 2017;45:12195–213.
Yue F, Cheng Y, Breschi A, Vierstra J, Wu W, Ryba T, et al. A comparative encyclopedia of DNA elements in the mouse genome. Nature.2014;515:355–64.
Kim MJ, Kim HS, Lee S, Min KY, Choi WS, You JS. Hexosamine biosynthetic pathway-derived O-GlcNAcylation is critical for RANKL-mediated osteoclast differentiation. Int J Mol Sci. 2021;22:8888.
Lonsdale J, Thomas J, Salvatore M, Phillips R, Lo E, Shad S, et al. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–5.
Zhu L, Li Q, Wong SH, Huang M, Klein BJ, Shen J, et al. ASH1L links Histone H3 lysine 36 dimethylation to MLL leukemia. Cancer Discov. 2016;6:770–83.
Yin B, Yu F, Wang C, Li B, Liu M, Ye L. Epigenetic control of mesenchymal stem cell fate decision via histone methyltransferase Ash1l. Stem Cells. 2019;37:115–27.
Mcgeary SE, Lin KS, Shi CY, Pham TM, Bisaria N, Kelley GM, et al. The biochemical basis of microRNA targeting efficacy. Science. 2019;366:eaav1741.
Colamaio M, Puca F, Ragozzino E, Gemei M, Decaussin-Petrucci M, Aiello C, et al. miR-142–3p down-regulation contributes to thyroid follicular tumorigenesis by targeting ASH1L and MLL1. J Clin Endocrinol Metab. 2015;100:E59–E69.
Aging Atlas Consortium. Aging Atlas: a multi-omics database for aging biology. Nucleic Acids Res. 2021;49:D825–D830.
Zheng Y, Liu X, Le W, Xie L, Li H, Wen W, et al. A human circulating immune cell landscape in aging and COVID-19. Protein Cell. 2020;11:740–70.
Liu YZ, Dvornyk V, Lu Y, Shen H, Lappe JM, Recker RR, et al. A novel pathophysiological mechanism for osteoporosis suggested by an in vivo gene expression study of circulating monocytes. J Biol Chem. 2005;280:29011–6.
Woetzel D, Huber R, Kupfer P, Pohlers D, Pfaff M, Driesch D, et al. Identification of rheumatoid arthritis and osteoarthritis patients by transcriptome-based rule set generation. Arthritis Res Ther. 2014;16:R84.
Chen X, Wang Z, Duan N, Zhu G, Schwarz EM, Xie C. Osteoblast-osteoclast interactions. Connect Tissue Res 2018;59:99–107.
Lee SH, Rho J, Jeong D, Sul JY, Kim T, Kim N, et al. v-ATPase V0 subunit d2-deficient mice exhibit impaired osteoclast fusion and increased bone formation. Nat Med. 2006;12:1403–9.
Negishi-Koga T, Shinohara M, Komatsu N, Bito H, Kodama T, Friedel RH, et al. Suppression of bone formation by osteoclastic expression of semaphorin 4D. Nat Med. 2011;17:1473–80.
Li D, Liu J, Guo B, Liang C, Dang L, Lu C, et al. Osteoclast-derived exosomal miR-214-3p inhibits osteoblastic bone formation. Nat Commun. 2016;7:10872.
Noble CL, Abbas AR, Cornelius J, Lees CW, Ho GT, Toy K, et al. Regional variation in gene expression in the healthy colon is dysregulated in ulcerative colitis. Gut.2008;57:1398–405.
Schuettengruber B, Bourbon HM, Di Croce L, Cavalli G. Genome regulation by polycomb and trithorax: 70 years and counting. Cell. 2017;171:34–57.
Perugorria MJ, Wilson CL, Zeybel M, Walsh M, Amin S, Robinson S, et al. Histone methyltransferase ASH1 orchestrates fibrogenic gene transcription during myofibroblast transdifferentiation. Hepatology.2012;56:1129–39.
Zhao X, Lin S, Li H, Si S, Wang Z. Myeloperoxidase controls bone turnover by suppressing osteoclast differentiation through modulating reactive oxygen species level. J Bone Miner Res. 2021;36:591–603.
Mabuchi Y, Houlihan DD, Akazawa C, Okano H, Matsuzaki Y. Prospective isolation of murine and human bone marrow mesenchymal stem cells based on surface markers. Stem Cells Int. 2013;2013:507301.
Shen J, Yang H, Qiao X, Chen Y, Zheng L, Lin J, et al. The E3 ubiquitin ligase TRIM17 promotes gastric cancer survival and progression via controlling BAX stability and antagonizing apoptosis. Cell Death Differ. 2023;30:2322–35.
Mei S, Qin Q, Wu Q, Sun H, Zheng R, Zang C, et al. Cistrome Data Browser: a data portal for ChIP-Seq and chromatin accessibility data in human and mouse. Nucleic Acids Res. 2017;45:D658–D662.
Bardou P, Mariette J, Escudié F, Djemiel C, Klopp C. jvenn: an interactive Venn diagram viewer. BMC Bioinform. 2014;15:293.
Funding
This work is supported by grants from the National Natural Science Foundation of China (No. 82100945) and the CAMS Innovation Fund for Medical Sciences (CIFMS) (Nos. 2021-I2M-1-055, 2021-I2M-1-030).
Author information
Authors and Affiliations
Contributions
Conception and design: ZW, XZ, SL; Development of methodology: XZ, SL, HR, SS, ZW; Acquisition of data: XZ, SL, HR, SS, LZ; Analysis and interpretation of data: XZ, SL, HR, ZW; Administrative, technical, or material support: ZW, LC. Writing of the manuscript: ZW, XZ, HR, LC. All the authors have authorized the submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics
All experimental procedures were ethically approved by the Institutional Animal Care and Use Committee of Institute of Medicinal Biotechnology, Chinese Academy of Medical Sciences & Peking Union Medical College.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhao, X., Lin, S., Ren, H. et al. The histone methyltransferase ASH1L protects against bone loss by inhibiting osteoclastogenesis. Cell Death Differ (2024). https://doi.org/10.1038/s41418-024-01274-w
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41418-024-01274-w