Abstract
Background
CA125 is the best available yet insufficiently sensitive biomarker for early detection of ovarian cancer. There is a need to identify novel biomarkers, which individually or in combination with CA125 can achieve adequate sensitivity and specificity for the detection of earlier-stage ovarian cancer.
Methods
In the European Prospective Investigation into Cancer and Nutrition (EPIC) cohort, we measured serum levels of 92 preselected proteins for 91 women who had blood sampled ≤18 months prior to ovarian cancer diagnosis, and 182 matched controls. We evaluated the discriminatory performance of the proteins as potential early diagnostic biomarkers of ovarian cancer.
Results
Nine of the 92 markers; CA125, HE4, FOLR1, KLK11, WISP1, MDK, CXCL13, MSLN and ADAM8 showed an area under the ROC curve (AUC) of ≥0.70 for discriminating between women diagnosed with ovarian cancer and women who remained cancer-free. All, except ADAM8, had shown at least equal discrimination in previous case-control comparisons. The discrimination of the biomarkers, however, was low for the lag-time of >9–18 months and paired combinations of CA125 with any of the 8 markers did not improve discrimination compared to CA125 alone.
Conclusion
Using pre-diagnostic serum samples, this study identified markers with good discrimination for the lag-time of 0–9 months. However, the discrimination was low in blood samples collected more than 9 months prior to diagnosis, and none of the markers showed major improvement in discrimination when added to CA125.
Similar content being viewed by others
Background
Current strategies for epithelial ovarian cancer (EOC) screening use a combination of blood-based biomarkers, notably cancer antigen 125 (CA125; mucin 16 (MUC16)) and human epididymis protein (HE)-4, and trans-vaginal ultrasound imaging. However, findings from randomised screening trials and prospective population cohorts have shown insufficient sensitivity and specificity of CA125 and HE4—the currently best two available markers—for the detection of early-stage ovarian tumours [1,2,3,4,5]. Thus, substantial effort is being directed to the search for additional protein biomarkers which either individually or in combination with CA125 and other markers could enhance the sensitivity and specificity for detecting ovarian cancer at an earlier, more treatable stage.
Olink® Proteomics has developed a technology based on the proximity extension assay (PEA) [6, 7], which permits the simultaneous measurement of up to 92 proteins in microliter volumes of blood serum or plasma. Several recent studies have used this multiplex platform to identify biomarkers for ovarian cancer detection, measuring candidate markers in blood samples collected from patients with clinically manifest ovarian cancer and from healthy controls or patients with benign pelvic conditions [8,9,10,11,12]. Using various (only partially overlapping) Olink® assay panels for sets of proteins relevant in oncology, inflammation and other disease areas, these studies identified several candidate proteins that, alone or in multi-marker panels, showed good discrimination between ovarian cancer patients and women with benign conditions or healthy controls. All studies, however, were based on classical case-control comparisons between serum or plasma samples from patients with clinically manifest (and mostly advanced-stage) ovarian cancer and cancer-free control subjects, and so far, there have been no studies examining these markers in blood samples collected from women prior to known cancer, and whether they may help increase the lead time for detection of ovarian cancer.
We here present findings from the first prospective study to evaluate the discriminatory performance of 92 oncology-related protein markers (Olink® Proseek Multiplex Oncology II panel) as potential early diagnostic biomarkers of ovarian cancer. We assessed the ability of the proteins to distinguish women having a future diagnosis of ovarian cancer from healthy controls using measurements from serum samples collected up to 18 months prior to diagnosis. Discrimination capacity is examined within strata of lag-time (0–9 months; >9–18 months) between blood draw and ovarian cancer diagnosis. For markers showing significant differences between case and controls, we corroborate findings through comparisons with the results from previous studies that used similar technology.
Study setting and methods
Case-control study, nested within the EPIC cohort
We conducted a case-control study nested within the European Prospective Investigation into Cancer and Nutrition (EPIC) cohort—a population-based, multi-center prospective cohort study in 10 European countries coordinated by the International Agency for Research on Cancer (IARC; Lyon, France) [13]. From 1992 to 2000, 366,521 women were enrolled and of these, 226,673 women provided a blood sample at baseline.
The present work is an extension of an earlier study on the prospective discriminatory capacity of CA125 and other early detection markers for ovarian cancer [4]. It includes pre-diagnostic serum samples from all incident cases (N = 91) of epithelial invasive ovarian (International Classification of Diseases for Oncology (ICD-O) code: C569), fallopian tube (C570) or peritoneal cancers (C480, C481, C482 and C488) with available data on tumour histology and diagnosed within maximally 18 months of blood draw (Table 1). All ovarian cancer cases were ascertained prospectively through record linkage with cancer and pathology registries (all countries except France, Germany, Greece, and Naples, Italy), or through active follow-up and systematic verification of self-reports by detailed examination and coding of clinical records (France, Germany and Naples, Italy). Information on tumour stage was available in part from pathology reports and in part from cancer registries, and for uniformity was coded into either local disease (stage I) or high-stage disease (regionally spread or metastatic). Information on tumour characteristics (histologic subtype [serous, endometrioid, clear cell, mucinous, not otherwise specified (NOS)]) and grade was additionally obtained from pathology reports.
For each of the 91 case subjects, two control participants (N = 182) were randomly selected among appropriate risk sets consisting of all female cohort members with a blood sample, alive and free of cancer at the time of diagnosis of the index case. An incidence density sampling protocol was used, such that, in principle, control participants could include women who became a cancer case later in time and each control participant could be sampled more than once; however, no control was actually drawn more than once and none of the control participants have subsequently been identified as ovarian cancer cases. Case and control participants were matched on study recruitment centre, age at blood draw (±6 months), time of the day of blood collection (±1 h), fasting status at blood collection, menopausal status at blood collection (premenopausal, perimenopausal, postmenopausal), current use of oral contraceptives or postmenopausal hormone replacements at the time of blood draw and phase of menstrual cycle for premenopausal women (3–5 categories; menstrual phase, follicular phase, ovulatory phase, luteal phase, or unknown, depending on available data).
Laboratory assays
The Proseek Multiplex Oncology II panel assays were performed in an Olink® certified laboratory at the German Center for Environmental Health (Helmholtz Zentrum München), Neuherberg, Germany. The proximity extension assay (PEA) technology, commercialised by Olink® Proteomics (Uppsala, Sweden), is a highly specific antibody-based technology that allows for relative quantification of numerous human protein biomarkers in body fluids [7]. Serum samples were analysed in batches, sorted by study centre and with samples from matched case-control sets together (in randomised and blinded order) in the same batch. The laboratory personnel were blinded regarding case-control status of the samples analysed. Results of the assays are reported in arbitrary units called ‘normalized protein expression’ values (NPX), which are relative protein expression levels from RT -qPCR on a log2 scale. For small proportions of study subjects, and for a few proteins, PEA measurements fell below the detection limit (IL6 [15%], FADD [26%)] CTSV [3%], MIC-A/B [2%] and CEACAM5 [18%]). When assay results were below the limit of detection, we assigned values to the midpoint between zero and the lower limit of detection.
Statistical analyses
We used unconditional logistic regression modelling for the estimation of covariate-adjusted receiver operating characteristic (ROC) curves, with calculation of area under curve (AUC) as an overall measure of the markers’ capacity to discriminate future cancer cases from participants who remained ovarian cancer-free. Models were systematically adjusted for study centre, age, menopausal status and use of hormone replacement therapy (HRT) at the time of blood draw as covariates. Analyses focused first on single markers. All models were fit for diagnosis of ovarian cancer of any histologic subtype as outcome and based on these models, individual ovarian cancer risk scores were calculated. The discrimination capacity of the overall risk scores was then further examined by strata of lag-time (≤9 months, >9–18 months) and by histologic subtype (serous vs non-serous or undetermined). Internal validation with 1000‐fold bootstrapping was used to adjust estimates of discriminative capacity for over‐optimism as a result of model overfitting. For all markers showing AUCs ≥0.70, we additionally estimated the sensitivity at 95% and at 98% specificity at cut-off points determined in our datasets for all women who remained cancer-free (N = 182).
For markers that showed discrimination of AUC ≥ 0.70 in the 0–9 months lag-time interval, and which had been highlighted as having discrimination potential in at least one previous study (Supplementary Table s1) based on clinical case-control comparisons, we further tested combined discrimination capacity jointly with CA125, using a two-marker discrimination model. These models were first fitted on the full dataset of 91 ovarian cancer cases and 182 controls, covering all lag-times from 0 to 18 months and risk scores were derived based on these models. The discrimination performance of the risk scores was then evaluated by strata of lag-time and histologic subtype. Likelihood-ratio tests were used to test whether the two-marker models significantly improved statistical model fit (and hence discrimination) compared to a model based on MUC16/CA125 only. We performed further analyses to examine the joint discrimination of two-marker model risk scores for tumours diagnosed within 0–9 or >9–18 months after blood draw, as well as by serous or other tumour histology, again using bootstrapping to adjust for over-optimism.
We also investigated whether in more exploratory approach that included all the 92 biomarkers irrespective of their univariate classification power, we could identify markers or panel of markers for discriminating between cases and women who remained cancer-free. This analysis included all women who remained cancer-free and those who were diagnosed with ovarian cancer 9–18 months after blood draw. It is among these women that the discrimination strength of CA125 relative to other marker candidates is not so dominant and improvements in discrimination are likely to be clinically relevant. As biomarkers are expected to be correlated, we used a least absolute shrinkage and selection operator (LASSO) algorithm implemented with the glmnet package in R [14] to select a parsimonious model for predicting ovarian cancer diagnosis. All analyses were conducted in SAS, version 9.4 (SAS Institute, Cary, NC) and R version 4.1.0 and RStudio version 1.4.1717 (The R Foundation for Statistical Computing).
Results
The characteristics of the ovarian cancer cases and matched controls are presented in Table 1. Of the 91 cases, 51 (56%) had serous tumours, whereas the remaining cases had tumours classified as mucinous (N = 10; 11%), endometroid (N = 7; 8%) clear cell (N = 4%) or tumours of non-specified type (NOS, N = 15; 16%). The majority (76.5%) of serous ovarian cancers were diagnosed at late stage (Stage III) while half (50%) of the non-serous cancers were at late stage. The distribution of disease spread and grade at diagnosis by histologic subtype is presented in Supplementary Table s2.
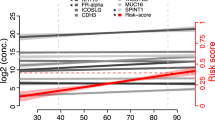
Nine biomarkers had an AUC of at least 0.7 for discriminating between women who developed ovarian cancer and women who remained ovarian cancer-free for at least 9 months after blood draw (Supplementary Table s3). The distribution of the levels of the nine markers in controls and in cases (by lag-time) are presented in Fig. 1. After performing 1000-fold bootstrapping to correct for potential over-optimism, all the nine markers maintained at least an AUC of 0.7 for distinguishing between controls and ovarian cancer cases diagnosed within 9 months of blood draw (Table 2). Of the 9 biomarkers, 8 had been previously reported to be informative for discrimination in at least one previous study comparing between ovarian cancer patients and healthy controls or patients with benign tumours (for an overview of the previous study findings, see Supplementary Table s1). CA125 (MUC16) and HE4 (WFDC2) had the highest discrimination with AUCs of 0.77 (95% CI: 0.75–0.79) and 0.73 (95% CI: 0.71–0.74), respectively, for the entire time lag of 0–18 months. The other markers that had been highlighted at least twice in previous studies and had good discrimination between healthy women and women who were diagnosed with either serous or non-serous ovarian cancer within 9 months of blood collection in the current study were: folate receptor alpha (FOLR1/FR-alpha), kallikrein (KLK11), midkine (MDK/MK) and C-X-C motif chemokine 13 (CXCL13). Two other markers—WNT1-inducible signaling pathway protein 1 (WISP1) and mesothelin (MSLN) had been found to have discriminatory potential in at least one cross-sectional case-control comparison (Table 2).
Each of the panels shows marker distributions in the form of Box plots, for biomarkers that yielded an AUC ≥ 0.7 for ovarian cancer diagnosis 0–9 months after blood draw. For the cancer cases, the plots show marker distributions measured in blood samples that had been collected 0–9 or >9–18 months prior to diagnosis.
All the nine best-performing biomarkers showed a decay in discrimination strength with longer lag-time between blood draw and ovarian cancer diagnosis (Table 2). For instance, there were five markers with AUCs of at least 0.7 for discriminating between healthy women and ovarian cancer cases (all histologies) for the lag-time of 0–9 months but only CA125 had an AUC higher than 0.7 for the lag-time of >9–18 months. As a second example, the discrimination performance of FR-alpha was 0.73 (95% CI: 0.69–0.77) for the lag-time of 0–9 months and only 0.62 (95% CI: 0.59–0.64) for the lag-time of 9–18 months. Similar reductions in discrimination performance with longer lag-time between blood draw and diagnosis were observed for all other markers. HE4/WFDC2 had an AUC of 0.63 (95% CI: 0.61–0.66) for the lag-time of 9–18 months (Table 2). Based on their sensitivities at 95% and at 98% specificity, the nine markers showed a similar decay in performance (Table 3). Within the lag period of 0–9 months, most markers showed stronger discrimination for serous tumours as compared to tumours with other or unspecified histology, with notable exceptions for WISP1, and CXCL13 which showed higher AUCs for the tumours of non-serous histology.
For all the eight markers that individually had AUCs ≥ 0.70, we examined the discrimination potential of the markers alone and in combination with CA125 (Table 4; Fig. 2). Using the likelihood-ratio test (LRT), we tested improvement in model fit of adding any of the eight markers to a model containing CA125 alone. None of the biomarkers resulted in significant improvements in model fit. We also observed only minor improvements in the corresponding joint discrimination with magnitude of 1%-point increase in discrimination strength compared to CA125 alone. Subtle improvements in discrimination were noted in further analysis by lag-time and histology. For instance, combination of CA125 and HE4 had a 1%-point higher performance than CA125 alone for all ovarian cancer histologies combined, for the lag-time of 0–9 months. Similarly, for the lag-time of >9–18 months, a combination of CA125 with ADAM8 had a slightly better performance, by 2% points than CA125 alone (Table 3). All the eight biomarkers were positively correlated with CA125 (Supplementary Fig. 1).
To further explore whether any marker combinations exited that might contribute discrimination information beyond that provided by CA125 alone, we conducted further analyses using a more exploratory approach including all 92 markers. The analyses employed a LASSO algorithm for model selection and included cases diagnosed 9–18 months following blood draw and all women who remained cancer-free. We found that the most regularised parsimonious model for predicting case-control status of ovarian cancer, at value of regularisation parameter (λ) such that the cross-validated error is within one standard error of the minimum, i.e. lambda.1se, contained only CA125.
Discussion
Using serum samples and data collected in the EPIC cohort, we examined the capacity of 92 cancer-related protein biomarkers measured using the Olink® Proteomics Oncology II panel to discriminate between women who were prospectively diagnosed with ovarian cancer and matched control women who remained cancer-free. In the analysis of women who developed ovarian cancer less than 9 months after blood draw, nine biomarkers showed potentially useful discrimination, with AUC ≥ 0.70. Besides the well-established markers CA125 and HE4, four other markers in this list: (FR-alpha, KLK11, MDK and CXCL13) had been highlighted previously as having discrimination potential in several prior case-control comparisons using the Olink® Multiplex platforms [8,9,10,11,12, 15], as well as in some further studies using other platforms [16,17,18]. We did not observe meaningful improvements in diagnostic performance by adding a single markers to CA125, particularly for ovarian cancer diagnosed more than 9 months after blood sampling.
Eight of the nine best-performing biomarkers had been previously reported by at least one case-control study to show good discrimination between ovarian cancer cases and controls. This concordance with earlier findings suggests that the proteins could be genuinely associated with cancer development and indeed, most of the biomarkers have been implicated to play a role in ovarian carcinogenesis or associated with ovarian cancer prognosis. FOLR1/FR-alpha is involved in the unidirectional transportation of folates into cells, metabolism of which facilitates DNA synthesis, methylation and repair [19]. In normal ovarian tissue, the expression of FOLR1 is restricted to luminal surfaces but is ubiquitous in ovarian tumour tissue, mostly in tumours of non-mucinous histology [20]. CXCL13 has been shown to play a role in immune cell recruitment to the site of chronic inflammation, activation and adaptive immune response regulation [21]. KLK11/hK11 was reported to be highly expressed in ovarian cancer patients, mostly early-stage tumours and is thus a potential marker of favourable prognosis [22]. WISP1 is believed to play a role in a number of cancers and is associated with poor survival and clinical grades of endometrial adenocarcinoma (endometrioid type) [23, 24].
The diagnostic performance of CA125, HE4 and other candidate biomarkers on the panel markedly reduced with increasing lag-time between blood draw and cancer diagnosis, which is in line with previous studies [3, 4, 25], and is to be expected for markers genuinely associated with tumour development. However, to be beneficial, markers or marker combinations should provide sufficient early detection lead time, such that earlier medical intervention can improve a patient’s survival. Previous studies suggested that, while at least for some patients serum biomarker levels indicative of ovarian cancer could be detectable at most 3 years prior to diagnosis, for most patients the likely lead time was less than 1 year [3, 4, 25,26,27]. We found that other than CA125, none of the other markers provided useful discriminatory information for ovarian cancer detected more than 9 months after blood draw and that paired combinations of CA125 with any other of the 8 markers did not meaningfully improve discrimination compared to CA125 alone, in either the 0–9 or the >9–18 months lag-time intervals. Similarly, in a more exploratory approach using LASSO algorithms that sought to identify markers or panel of markers for discriminating between cases and controls in samples collected 9–18 months prior to diagnosis, and regardless of the univariate classification power of the biomarker or being previously shown to have discrimination potential, we still could not identify markers that improved on the discrimination of CA125, without model overfitting. Thus, given that CA125 is the best available but insufficiently sensitive marker of ovarian cancer, our findings suggest that none of the biomarkers investigated in this study has sufficient potential to extend the lead time longer than that provided by CA125 alone.
Our study has some limitations. For women who were diagnosed with ovarian cancer, we have no knowledge about the stage of the tumours at the time of blood draw, making it difficult to speculate whether improved discrimination performance of the markers would result in survival benefit. Also, as we examined a total of 92 markers, our analyses may have resulted in false-positive leads observed only by chance, although most of the markers with AUC > 0.7 (0–9-month interval) in our present dataset had shown discrimination potential in previous case-control studies including prevalent cases. Conversely, due to the limited sample size it is possible that some markers truly associated with ovarian cancer were missed. When testing two-marker combinations (CA125 plus any other marker) we observed no meaningful improvements in discrimination. Developing accurately weighted marker scores for more than two markers (more than one additionally to CA125) will require larger numbers of ovarian cancer cases than in our present study and could be achieved by combining serum samples and data of ovarian cancer cases and matched control subjects from additional large-scale population cohorts worldwide.
Conclusion
Our study confirms the good discrimination between ovarian cancer cases and controls of several biomarkers previously observed in cross-sectional studies. However, markers showed discrimination only in samples collected 9 months prior to ovarian cancer diagnosis and much less so in samples collected 9–18 months prior to diagnosis. Unfortunately, combining single markers with CA125 did not improve the diagnostic performance of the markers.
Disclaimer
Where authors are identified as personnel of the International Agency for Research on Cancer/World Health Organization, the authors alone are responsible for the views expressed in this article and they do not necessarily represent the decisions, policy or views of the International Agency for Research on Cancer/World Health Organization.
Data availability
The EPIC project was launched in the 1990s. Unlike in new studies that we run today, public access to data from the EPIC population was not part of the study protocol at that time. Thus, the data protection statement and informed consent of the EPIC participants do not cover the provision of data in public repositories. Nevertheless, we are open to provide our dataset upon request for (a) statistical validation by reviewers and (b) pooling projects under clearly defined and secure conditions and based on valid data transfer agreements.
References
Jacobs IJ, Menon U, Ryan A, Gentry-Maharaj A, Burnell M, Kalsi JK, et al. Ovarian cancer screening and mortality in the UK Collaborative Trial of Ovarian Cancer Screening (UKCTOCS): a randomised controlled trial. Lancet Lond Engl. 2016;387:945–56.
Cohen JG, White M, Cruz A, Farias-Eisner R. In 2014, can we do better than CA125 in the early detection of ovarian cancer? World J Biol Chem. 2014;5:286–300.
Cramer DW, Bast RC, Berg CD, Diamandis EP, Godwin AK, Hartge P, et al. Ovarian cancer biomarker performance in prostate, lung, colorectal, and ovarian cancer screening trial specimens. Cancer Prev Res. 2011;4:365–74.
Terry KL, Schock H, Fortner RT, Hüsing A, Fichorova RN, Yamamoto HS, et al. A prospective evaluation of early detection biomarkers for ovarian cancer in the European EPIC cohort. Clin Cancer Res J Am Assoc Cancer Res. 2016;22:4664–75.
Menon U, Gentry-Maharaj A, Burnell M, Singh N, Ryan A, Karpinskyj C, et al. Ovarian cancer population screening and mortality after long-term follow-up in the UK Collaborative Trial of Ovarian Cancer Screening (UKCTOCS): a randomised controlled trial. Lancet Lond Engl. 2021;397:2182–93.
Lundberg M, Eriksson A, Tran B, Assarsson E, Fredriksson S. Homogeneous antibody-based proximity extension assays provide sensitive and specific detection of low-abundant proteins in human blood. Nucleic Acids Res. 2011;39:e102.
Assarsson E, Lundberg M, Holmquist G, Björkesten J, Thorsen SB, Ekman D, et al. Homogenous 96-plex PEA immunoassay exhibiting high sensitivity, specificity, and excellent scalability. PLoS ONE. 2014;9:e95192.
Leandersson P, Åkesson A, Hedenfalk I, Malander S, Borgfeldt C. A multiplex biomarker assay improves the diagnostic performance of HE4 and CA125 in ovarian tumor patients. PLoS ONE. 2020;15:e0240418.
Boylan KLM, Geschwind K, Koopmeiners JS, Geller MA, Starr TK, Skubitz APN. A multiplex platform for the identification of ovarian cancer biomarkers. Clin Proteomics. 2017;14:34.
Skubitz APN, Boylan KLM, Geschwind K, Cao Q, Starr TK, Geller MA, et al. Simultaneous measurement of 92 serum protein biomarkers for the development of a multiprotein classifier for ovarian cancer detection. Cancer Prev Res. 2019;12:171–84.
Enroth S, Berggrund M, Lycke M, Lundberg M, Assarsson E, Olovsson M, et al. A two-step strategy for identification of plasma protein biomarkers for endometrial and ovarian cancer. Clin Proteomics. 2018;15:38.
Enroth S, Berggrund M, Lycke M, Broberg J, Lundberg M, Assarsson E, et al. High throughput proteomics identifies a high-accuracy 11 plasma protein biomarker signature for ovarian cancer. Commun Biol. 2019;2:1–12.
Riboli E, Hunt KJ, Slimani N, Ferrari P, Norat T, Fahey M, et al. European prospective investigation into cancer and nutrition (EPIC): study populations and data collection. Public Health Nutr. 2002;5:1113–24.
Friedman JH, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw. 2010;33:1–22.
Graumann J, Finkernagel F, Reinartz S, Stief T, Brödje D, Renz H, et al. Multi-platform affinity proteomics identify proteins linked to metastasis and immune suppression in ovarian cancer plasma. Front Oncol. 2019. https://doi.org/10.3389/fonc.2019.01150.
Rice GE, Edgell TA, Autelitano DJ. Evaluation of midkine and anterior gradient 2 in a multimarker panel for the detection of ovarian cancer. J Exp Clin Cancer Res. 2010;29:62.
Derycke MS, Pambuccian SE, Gilks CB, Kalloger SE, Ghidouche A, Lopez M, et al. Nectin 4 overexpression in ovarian cancer tissues and serum: potential role as a serum biomarker. Am J Clin Pathol. 2010;134:835–45.
Leung F, Bernardini MQ, Brown MD, Zheng Y, Molina R, Bast RC, et al. Validation of a novel biomarker panel for the detection of ovarian cancer. Cancer Epidemiol Biomarkers Prev. 2016;25:1333–40.
Elnakat H, Ratnam M. Role of folate receptor genes in reproduction and related cancers. Front Biosci J Virtual Libr. 2006;11:506–19.
Kalli KR, Oberg AL, Keeney GL, Christianson TJH, Low PS, Knutson KL, et al. Folate receptor alpha as a tumor target in epithelial ovarian cancer. Gynecol Oncol. 2008;108:619–26.
Rao DA. T cells that help B cells in chronically inflamed tissues. Front Immunol. 2018. https://doi.org/10.3389/fimmu.2018.01924.
Diamandis EP, Borgoño CA, Scorilas A, Harbeck N, Dorn J, Schmitt M. Human kallikrein 11: an indicator of favorable prognosis in ovarian cancer patients. Clin Biochem. 2004;37:823–9.
Liu Y, Song Y, Ye M, Hu X, Wang ZP, Zhu X. The emerging role of WISP proteins in tumorigenesis and cancer therapy. J Transl Med. 2019;17:28.
Tang Q, Jiang X, Li H, Lin Z, Zhou X, Luo X, et al. Expression and prognostic value of WISP-1 in patients with endometrial endometrioid adenocarcinoma. J Obstet Gynaecol Res. 2011;37:606–12.
Anderson GL, McIntosh M, Wu L, Barnett M, Goodman G, Thorpe JD, et al. Assessing lead time of selected ovarian cancer biomarkers: a nested case-control study. J Natl Cancer Inst. 2010;102:26–38.
Gentry-Maharaj A, Blyuss O, Ryan A, Burnell M, Karpinskyj C, Gunu R, et al. Multi-marker longitudinal algorithms incorporating HE4 and CA125 in ovarian cancer screening of postmenopausal women. Cancers (Basel). 2020;12:1931. https://doi.org/10.3390/cancers12071931.
Simmons AR, Fourkala EO, Gentry-Maharaj A, Ryan A, Sutton MN, Baggerly K, et al. Complementary longitudinal serum biomarkers to CA125 for early detection of ovarian cancer. Cancer Prev Res. 2019;12:391–400.
Acknowledgements
We acknowledge the use of data and biological samples from the EPIC-Asturias cohort, PI Ramón Quirós, the EPIC-Bilthoven cohort, PI W. M. Monique Verschuren, the EPIC-France cohort, PI Gianluca Severi, the EPIC- Utrecht, PI Roel Vermeulen and the EPIC-Norfolk cohort, PI Nick Wareham. We thank the Julius Center for Health Sciences and Primary Care, University Medical Center Utrecht, Utrecht University, Utrecht, the Netherlands and the National Institute for Public Health and the Environment (RIVM), Bilthoven, the Netherlands, for their contribution and ongoing support to the EPIC Study. We are grateful for the continuous participation of our cohort participants—without their commitment, this work would not have been possible.
Funding
The coordination of EPIC is financially supported by International Agency for Research on Cancer (IARC) and also by the Department of Epidemiology and Biostatistics, School of Public Health, Imperial College London-, which has additional infrastructure support provided by the NIHR Imperial Biomedical Research Centre (BRC). The national cohorts are supported by: Danish Cancer Society (Denmark); Ligue Contre le Cancer, Institut Gustave Roussy, Mutuelle Générale de l’Education Nationale, Institut National de la Santé et de la Recherche Médicale (INSERM) (France); German Cancer Aid, German Cancer Research Center (DKFZ), German Institute of Human Nutrition Potsdam-Rehbruecke (DIfE), Federal Ministry of Education and Research (BMBF) (Germany); Associazione Italiana per la Ricerca sul Cancro-AIRC-Italy, Compagnia di SanPaolo and National Research Council (Italy); Dutch Ministry of Public Health, Welfare and Sports (VWS), Netherlands Cancer Registry (NKR), LK Research Funds, Dutch Prevention Funds, Dutch ZON (Zorg Onderzoek Nederland), World Cancer Research Fund (WCRF), Statistics Netherlands (The Netherlands); Health Research Fund (FIS)—Instituto de Salud Carlos III (ISCIII), Regional Governments of Andalucía, Asturias, Basque Country, Murcia and Navarra,- and the Catalan Institute of Oncology—ICO (Spain); Swedish Cancer Society, Swedish Research Council and County Councils of Skåne and Västerbotten (Sweden); Cancer Research UK (C864/A14136 to EPIC-Norfolk; C8221/A29017 to EPIC-Oxford), Medical Research Council (MR/N003284/1, MC-UU 12015/1 and MC UU_00006/1to EPIC-Norfolk; MR/M012190/1 to EPIC-Oxford). (United Kingdom). Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
TM: Formal analysis, resources, visualisation, investigation, methodology, writing–original draft, writing—review and editing. RTF: Conceptualisation, resources, formal analysis, funding acquisition, investigation, methodology, writing—original draft, writing—review and editing. VK: Conceptualisation, resources, investigation, methodology, writing—review and editing. LCH: Formal analysis, investigation, methodology, writing—review and editing. AP: Resources, investigation, methodology, writing—review and editing. SMH: Resources, investigation, methodology, writing—review and editing. TJ: Conceptualisation, resources, methodology, writing—original draft, writing—review and editing. MS: Resources, funding acquisition, writing—review and editing. CS: Resources, funding acquisition, writing—review and editing. ALRH: Resources, funding acquisition, writing—review and editing. AT: Resources, funding acquisition, writing—review and editing. KO: Resources, funding acquisition, writing—review and editing. MJSP: Resources, funding acquisition, writing—review and editing. MCB: Resources, funding acquisition, writing—review and editing. MDC: Resources, funding acquisition, writing—review and editing. PA: Resources, funding acquisition, writing—review and editing. EA: Resources, funding acquisition, writing—review and editing. ELW: Resources, funding acquisition, writing—review and editing. RCT: Resources, funding acquisition, writing—review and editing. RC: Resources, funding acquisition, writing—review and editing. SG: Resources, funding acquisition, writing—review and editing. GM: Resources, funding acquisition, writing—review and editing. SS: Resources, funding acquisition, writing—review and editing. RT: Resources, funding acquisition, writing—review and editing. ITG: Resources, funding acquisition, writing—review and editing. TMS: Resources, funding acquisition, writing—review and editing. HS: Resources, funding acquisition, writing—review and editing. EL: Resources, funding acquisition, writing—review and editing. AI: Resources, funding acquisition, writing—review and editing. AKH: Resources, funding acquisition, writing—review and editing. LD: Resources, funding acquisition, writing—review and editing. EW: Resources, funding acquisition, writing—review and editing. RK: Conceptualisation, resources, formal analysis, funding acquisition, investigation, methodology, writing—original draft, writing—review and editing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
All EPIC study participants have given written consent for future analyses of their blood samples for research purposes. This study was approved by the International Agency for Research on Cancer (IARC, Lyon, France) Ethics Committee. The study was performed in accordance to the principles expressed in the Helsinki Declaration of 1996, conventions of the Council of Europe on Human Rights and Biomedicine, and the UNESCO Declaration on Human Genome.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Mukama, T., Fortner, R.T., Katzke, V. et al. Prospective evaluation of 92 serum protein biomarkers for early detection of ovarian cancer. Br J Cancer 126, 1301–1309 (2022). https://doi.org/10.1038/s41416-021-01697-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41416-021-01697-z
This article is cited by
-
Transcriptome analysis reveals the clinical significance of CXCL13 in Pan-Gyn tumors
Journal of Cancer Research and Clinical Oncology (2024)
-
Cytokine profiling identifies circulating IL-6 and IL-15 as prognostic stratifiers in patients with non-small cell lung cancer receiving anti-PD-1/PD-L1 blockade therapy
Cancer Immunology, Immunotherapy (2023)
-
Serum immune modulators associated with immune-related toxicities and efficacy of atezolizumab in patients with non-small cell lung cancer
Journal of Cancer Research and Clinical Oncology (2023)
-
Detection of plasma exosomal miRNA-205 as a biomarker for early diagnosis and an adjuvant indicator of ovarian cancer staging
Journal of Ovarian Research (2022)