Abstract
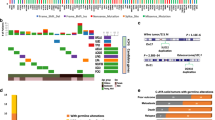
Imatinib represents the standard therapy for gastrointestinal stromal tumor (GIST) patients with metastatic/unresectable disease. Despite the excellent results achieved with its introduction, the majority of patients quite invariably experience disease progression. The aim of this study was to understand the contribution of germline DNA polymorphisms in discriminating between imatinib clinical response [evaluated as progression free survival (PFS)] and toxicity. In particular, a discovery cohort (34 GIST with a KIT exon 11 primary mutation, and no toxicity) was analyzed through DMET array that interrogates 1936 variants in 231 genes of the ADME process. We further confirmed the genotype of selected variants in an extended cohort of 49 patients (the original cohort and 15 new cases, all with exon 11 primary mutation), identifying 6 SNPs— ABCB4 rs1202283, ABCC2 rs2273697, ABCG1 rs1541290, CYP11B1 rs7003319, CYP7B1 rs6987861, and NQO1 rs10517—significantly associated with response to imatinib. Three SNPs, ABCB4 rs1202283, ABCC2 rs2273697, and NQO1 rs10517, which had a significant association after adjusted multivariate analysis, were included in a genetic prediction model. We confirmed that these SNPs could stratify the cohort of 49 patients according to the risk of developing progression under imatinib treatment. In conclusion, we identified a genetic signature of response to imatinib therapy in GIST patients able to stratify patients at low and high risk to progress, according to their genotype.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 6 print issues and online access
$259.00 per year
only $43.17 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Antonescu CR. The GIST paradigm: lessons for other kinase-driven cancers. J Pathol. 2011;223:251–61.
Corless CL, Barnett CM, Heinrich MC. Gastrointestinal stromal tumours: origin and molecular oncology. Nat Rev Cancer. 2011;11:865–78.
Heinrich MC, Corless CL, Duensing A, McGreevey L, Joseph N, Singer S, et al. PDGFRA activating mutations in gastrointestinal stromal tumors. Science. 2003;299:708–10.
Angelini S, Ravegnini G, Fletcher JA, Maffei F, Hrelia P. Clinical relevance of pharmacogenetics in gastrointestinal stromal tumor treatment in the era of personalized therapy. Pharmacogenomics. 2013;14:941–56.
Demetri GD, von Mehren M, Blanke CD, Van den Abbeele AD, Eisenberg B, Roberts PJ, et al. Efficacy and safety of imatinib mesylate in advanced gastrointestinal stromal tumors. N Engl J Med. 2002;347:472–80.
Demetri GD, van Oosterom AT, Garrett CR, Blachstein ME, Shah MH, Verweij J, et al. Efficacy and safety of sunitinib in patients with advanced gastrointestinal stromal tumour after failure of imatinib: a randomised controlled trial. Lancet. 2006;368:1329–38.
Heinrich MC, Corless CL, Demetri GD, Blanke CD, von Mehren M, Joensuu H, et al. Kinase mutations and imatinib response in patients with metastatic gastrointestinal stromal tumor. J Clin Oncol. 2003;21:4342–9.
Maleddu A, Pantaleo MA, Nannini M, Biasco G. The role of mutational analysis of KIT and PDGFRA in gastrointestinal stromal tumors in a clinical setting. J Transl Med. 2011;9:75.
Heinrich MC, Owzar K, Corless CL, Hollis D, Borden EC, Fletcher CD, et al. Correlation of kinase genotype and clinical outcome in the North American Intergroup Phase III Trial of imatinib mesylate for treatment of advanced gastrointestinal stromal tumor: CALGB 150105 Study by Cancer and Leukemia Group B and Southwest Oncology Group. J Clin Oncol. 2008;26:5360–7.
Debiec-Rychter M, Dumez H, Judson I, Wasag B, Verweij J, Brown M, et al. Use of c-KIT/PDGFRA mutational analysis to predict the clinical response to imatinib in patients with advanced gastrointestinal stromal tumours entered on phase I and II studies of the EORTC Soft Tissue and Bone Sarcoma Group. Eur J Cancer. 2004;40:689–95.
Gastrointestinal Stromal Tumor Meta-Analysis Group (MetaGIST). Comparison of two doses of imatinib for the treatment of unresectable or metastatic gastrointestinal stromal tumors: a meta-analysis of 1,640 patients. J Clin Oncol. 2010;28:1247–53.
Angelini S, Pantaleo MA, Ravegnini G, Zenesini C, Cavrini G, Nannini M, et al. Polymorphisms in OCTN1 and OCTN2 transporters genes are associated with prolonged time to progression in unresectable gastrointestinal stromal tumours treated with imatinib therapy. Pharmacol Res. 2013;68:1–6.
Serrano C, Wang Y, Mariño-Enríquez A, Lee JC, Ravegnini G, Morgan JA, et al. KRAS and KIT Gatekeeper Mutations Confer Polyclonal Primary Imatinib Resistance in GI Stromal Tumors: Relevance of Concomitant Phosphatidylinositol 3-Kinase/AKT Dysregulation. J Clin Oncol. 2015;33:e93–6.
Ravegnini G, Sammarini G, Angelini S, Hrelia P. Pharmacogenetics of tyrosine kinase inhibitors in gastrointestinal stromal tumor and chronic myeloid leukemia. Expert Opin Drug Metab Toxicol. 2016;12:733–42.
Debiec-Rychter M, Sciot R, Le Cesne A, Schlemmer M, Hohenberger P, van Oosterom AT, et al. KIT mutations and dose selection for imatinib in patients with advanced gastrointestinal stromal tumours. Eur J Cancer. 2006;42:1093–103.
Duensing A, Medeiros F, McConarty B, Joseph NE, Panigrahy D, Singer S, et al. Mechanisms of oncogenic KIT signal transduction in primary gastrointestinal stromal tumours (GISTs). Oncogene. 2004;23:3999–4006.
Ravegnini G, Nannini M, Sammarini G, Astolfi A, Biasco G, Pantaleo MA, et al. Personalized Medicine in Gastrointestinal Stromal Tumor (GIST): Clinical Implications of the Somatic and Germline DNA Analysis. Int J Mol Sci. 2015;16:15592–608.
Peng B, Lloyd P, Schran H. Clinical pharmacokinetics of imatinibby. Clin Pharmacokinet. 2005;44:879–94.
Oostendorp RL, Buckle T, Beijnen JH, van Tellingen O, Schellens JH. The effect of P-gp (Mdr1a/1b), BCRP (Bcrp1) and P-gp/BCRP inhibitors on the in vivo absorption, distribution, metabolism and excretion of imatinib. Invest New Drugs. 2009;27:31–40.
O’Brien KM, Orlow I, Antonescu CR, Ballman K, McCall L, DeMatteo R, et al. Gastrointestinal stromal tumors, somatic mutations and candidate genetic risk variants. PLoS ONE. 2013;8:e62119.
Koo DH, Ryu MH, Ryoo BY, Beck MY, Na YS, Shin JG, et al. Association of ABCG2 polymorphism with clinical efficacy of imatinib in patients with gastrointestinal stromal tumor. Cancer Chemother Pharmacol. 2015;75:173–82.
Angelini S, Ravegnini G, Nannini M, Bermejo JL, Musti M, Pataleo MA, et al. Folate-related polymorphisms in gastrointestinal stromal tumours: susceptibility and correlation with tumour characteristics and clinical outcome. Eur J Hum Genet. 2015;23:817–23.
Ravegnini G, Nannini M, Simeon V, Musti M, Sammarini G, Saponare M, et al. Polymorphisms in DNA repair genes in gastrointestinal stromal tumours: susceptibility and correlation with tumour characteristics and clinical outcome. Tumour Biol. 2016;37:13413–23.
Ravegnini G, Nannini M, Zenesini C, Simeon V, Sammarini G, Urbini M, et al. An exploratory association of polymorphisms in angiogenesis-related genes with susceptibility, clinical response and toxicity in gastrointestinal stromal tumors receiving sunitinib after imatinib failure. Angiogenesis. 2017;20:139–48.
Deeken JF, Cormier T, Price DK, Sissung TM, Steinberg SM, Tran K, et al. A pharmacogenetic study of docetaxel and thalidomide in patients with castration-resistant prostate cancer using the DMET genotyping platform. Pharm J. 2010;10:191–9.
Di Martino MT, Arbitrio M, Guzzi PH, Leone E, Baudi F, Piro E, et al. A peroxisome proliferator-activated receptor gamma (PPARG) polymorphism is associated with zoledronic acid-related osteonecrosis of the jaw in multiple myeloma patients: analysis by DMET microarray profiling. Br J Haematol. 2011;154:529–33.
Di Martino MT, Arbitrio M, Leone E, Guzzi PH, Rotundo MS, Ciliberto D, et al. Single nucleotide polymorphisms of ABCC5 and ABCG1 transporter genes correlate to irinotecan-associated gastrointestinal toxicity in colorectal cancer patients: a DMET microarray profiling study. Cancer Biol Ther. 2011;12:780–7.
Rumiato E, Boldrin E, Amadori A, Saggioro D. DMETTM (Drug-Metabolizing Enzymes and Transporters) microarray analysis of colorectal cancer patients with severe 5-fluorouracil-induced toxicity. Cancer Chemother Pharmacol. 2013;72:483–8.
Lombardi G, Rumiato E, Bertorelle R, Saggioro D, Farina P, Della Puppa A, et al. Clinical and genetic factors associated with severe hematological toxicity in glioblastoma patients during radiation plus temozolomide treatment: A Prospective Study. Am J Clin Oncol. 2015;38:514–9.
Nieuweboer AJ, Smid M, de Graan AJ, Elbouazzaoui S, de Bruijn P, Martens JW, et al. Predicting paclitaxel-induced neutropenia using the DMET platform. Pharmacogenomics. 2015;16:1231–41.
Arbitrio M, Di Martino MT, Barbieri V, Agapito G, Guzzi PH, Botta C, et al. Identification of polymorphic variants associated with erlotinib-related skin toxicity in advanced non-small cell lung cancer patients by DMET microarray analysis. Cancer Chemother Pharmacol. 2016;77:205–9.
Rumiato E, Boldrin E, Malacrida S, Battaglia G, Bocus P, Castoro C, et al. A germline predictive signature of response to platinum chemotherapy in esophageal cancer. Transl Res. 2016;171:29–37.e1.
Drenberg CD, Paugh SW, Pounds SB, Shi L, Orwick SJ, Li L, et al. Inherited variation in OATP1B1 is associated with treatment outcome in acute myeloid leukemia. Clin Pharmacol Ther. 2016;99:651–60.
Di Martino MT, Scionti F, Sestito S, Nicoletti A, Arbitrio M, Hiram Guzzi P, et al. Genetic variants associated with gastrointestinal symptoms in Fabry disease. Oncotarget. 2016;7:85895–904.
Arbitrio M, Di Martino MT, Scionti F, Agapito G, Guzzi PH, Cannataro M, et al. DMETTM (Drug Metabolism Enzymes and Transporters): a pharmacogenomic platform for precision medicine. Oncotarget. 2016;7:54028–50.
Agapito G, Guzzi PH, Cannataro M. DMET-Miner: efficinet discovery of association rules from pharmacogenomic data. J Biomed Inform. 2015;56:273–83.
Dumaual C, Miao X, Daly TM, bruckner C, Njau R, Fu DJ, et al. Comprehensive assessment of metabolic enzyme and transporter genes using the Affymetrix Targeted Genotyping System. Pharmacogenomics. 2007;8:293–305.
Guzzi PH, Agapito G, Di Martino MT, Arbitrio M, Tassone P, Tagliaferri P, et al. DMET-analyzer: automatic analysis of Affymetrix DMET data. BMC Bioinforma. 2012;13:258.
Au A, Baba AA, Azlan H, Norsa’adah B, Ankathil R. Clinical impact of ABCC1 and ABCC2 genotypes and haplotypes in mediating imatinib resistance among chronic myeloid leukaemia patients. J Clin Pharm Ther. 2014;39:685–90.
Chen Z, Shi T, Zhang L, Zhu P, Deng M, Huang C, et al. Mammalian drug efflux transporters of the ATP binding cassette (ABC) family in multidrug resistance: A review of the past decade. Cancer Lett. 2016;370:153–64.
Winger JA, Hantschel O, Superti-Furga G, Kuriyan J. The structure of the leukemia drug imatinib bound to human quinone reductase 2 (NQO2). BMC Struct Bio. 2009;9:7.
Chen F, Liu Y, Wang S, Guo X, Shi P, Wang W, et al. Triptolide, a Chinese herbal extract, enhances drug sensitivity of resistant myeloid leukemia cell lines through downregulation of HIF-1α and Nrf2. Pharmacogenomics. 2013;14:1305–17.
Ma D, Fang Q, Wang P, Gao R, Wu W, Lu T, et al. Induction of heme oxygenase-1 by Na + -H + exchanger 1 protein plays a crucial role in imatinib-resistant chronic myeloid leukemia cells. J Biol Chem. 2015;290:12558–71.
Synowiec E, Hoser G, Wojcik K, Pawlowska E, Skorski T, Blasiak J. UV Differentially Induces Oxidative Stress, DNA Damage and Apoptosis in BCR-ABL1-Positive Cells Sensitive and Resistant to Imatinib. Int J Mol Sci. 2015;16:18111–28.
Kim TW, Kim YJ, Kim HT, Park SR, Lee MY, Park YD, et al. NQO1 Deficiency Leads Enhanced Autophagy in Cisplatin-Induced Acute Kidney Injury Through the AMPK/TSC2/mTOR Signaling Pathway. Antioxid Redox Signal. 2016;24:867–83.
Ravegnini G, Sammarini G, Nannini M, Pantaleo MA, Biasco G, Hrelia P, et al. Gastrointestinal stromal tumors (GIST): Facing cell death between autophagy and apoptosis. Autophagy. 2017;13:452–63.
Acknowledgements
Gloria Ravegnini is supported by a MSD Italia fellowship granted by and on behalf of Merck Sharp & Dohme Corporation.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Ravegnini, G., Urbini, M., Simeon, V. et al. An exploratory study by DMET array identifies a germline signature associated with imatinib response in gastrointestinal stromal tumor. Pharmacogenomics J 19, 390–400 (2019). https://doi.org/10.1038/s41397-018-0050-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41397-018-0050-4
This article is cited by
-
The GIST of it all: management of gastrointestinal stromal tumors (GIST) from the first steps to tailored therapy. A bibliometric analysis
Langenbeck's Archives of Surgery (2024)
-
Pharmacogenomics and functional imaging to predict irinotecan pharmacokinetics and pharmacodynamics: the predict IR study
Cancer Chemotherapy and Pharmacology (2021)
-
HIF-1α regulates cellular metabolism, and Imatinib resistance by targeting phosphogluconate dehydrogenase in gastrointestinal stromal tumors
Cell Death & Disease (2020)