Abstract
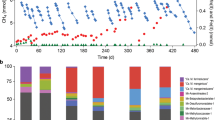
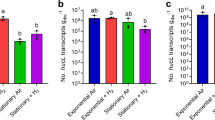
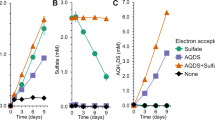
Although microbial humus respiration plays a critical role in organic matter decomposition and biogeochemical cycling of elements in diverse anoxic environments, the role of methane-producing species (methanogens) is not well defined. Here we report that a major fraction of humus, humic acid reduction enhanced the growth of Methanosarcina acetivorans above that attributed to methanogenesis when utilizing the energy sources methanol or acetate, results which showed both respiratory and fermentative modes of energy conservation. Growth characteristics with methanol were the same for an identically cultured mutant deleted for the gene encoding a multi-heme cytochrome c (MmcA), results indicating MmcA is not essential for respiratory electron transport to humic acid. Transcriptomic analyses revealed that growth with humic acid promoted the upregulation of genes annotated as cell surface pyrroloquinoline quinone (PQQ)-binding proteins. Furthermore, PQQ isolated from the membrane fraction was more abundant in humic acid-respiring cells, and the addition of PQQ improved efficiency of the extracellular electron transport. Given that the PQQ-binding proteins are widely distributed in methanogens, the findings extend current understanding of microbial humus respiration in the context of global methane dynamics.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
We are sorry, but there is no personal subscription option available for your country.
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Illumina sequence reads have been submitted to the SRA NCBI database under Bioprojects PRJNA859665 and PRJNA862363 including Biosamples SAMN29793682 (SRX16315150 to SRX16315152), SAMN29793683 (SRX16315153 to SRX16315155), SAMN29975242 (SRX16673125 to SRX16673127), and SAMN29975243 (SRX16673128 to SRX16673130).
References
Hayes M. Humus chemistry - genesis, composition, reactions - Stevenson,Fj. Nature. 1983;303:835–6.
Wang SQ, Wang YX, He XS, Lu QH. Degradation or humification: rethinking strategies to attenuate organic pollutants. Trends Biotechnol. 2022;40:1061–72.
Lipczynska-Kochany E. Humic substances, their microbial interactions and effects on biological transformations of organic pollutants in water and soil: a review. Chemosphere. 2018;202:420–37.
Lovley DR, Coates JD, BluntHarris EL, Phillips EJP, Woodward JC. Humic substances as electron acceptors for microbial respiration. Nature. 1996;382:445–8.
Lovley DR, Fraga JL, Blunt-Harris EL, Hayes LA, Phillips EJP, Coates JD. Humic substances as a mediator for microbially catalyzed metal reduction. Acta Hydroch Hydrob. 1998;26:152–7.
Scott DT, McKnight DM, Blunt-Harris EL, Kolesar SE, Lovely DR. Quinone moieties act as electron acceptors in the reduction of humic substances by humics-reducing microorganisms. Environ Sci Technol. 1999;32:2984–89.
Shi L, Dong HL, Reguera G, Beyenal H, Lu AH, Liu J, et al. Extracellular electron transfer mechanisms between microorganisms and minerals. Nat Rev Microbiol. 2016;14:651–62.
Roden EE, Kappler A, Bauer I, Jiang J, Paul A, Stoesser R, et al. Extracellular electron transfer through microbial reduction of solid-phase humic substances. Nat Geosci. 2010;3:417–21.
Klupfel L, Piepenbrock A, Kappler A, Sander M. Humic substances as fully regenerable electron acceptors in recurrently anoxic environments. Nat Geosci. 2014;7:195–200.
Valenzuela EI, Cervantes FJ. The role of humic substances in mitigating greenhouse gases emissions: current knowledge and research gaps. Sci Total Environ. 2021;750:141677.
Ferry JG. How to make a living by exhaling methane. Annu Rev Microbiol. 2010;64:453–73.
Prakash D, Chauhan SS, Ferry JG. Life on the thermodynamic edge: respiratory growth of an acetotrophic methanogen. Sci Adv. 2019;5:eaaw9059.
Yang Z, Lu Y. Coupling methanogenesis with iron reduction by acetotrophic Methanosarcina mazei zm-15. Environ Microbiol Rep. 2022;14:804–11.
Holmes DE, Ueki T, Tang HY, Zhou JJ, Smith JA, Chaput G, et al. A membrane-bound cytochrome enables methanosarcina acetivorans to conserve energy from extracellular electron transfer. mBio. 2019;10:e00789–19.
Welte C, Deppenmeier U. Bioenergetics and anaerobic respiratory chains of aceticlastic methanogens. BBA-Bioenerg. 2014;1837:1130–47.
Ferry JG. Methanosarcina acetivorans: a model for mechanistic understanding of aceticlastic and reverse methanogenesis. Front Microbiol. 2020;11:01806.
Gao KL, Lu YH. Putative extracellular electron transfer in methanogenic archaea. Front Microbiol. 2021;12:611739.
Duine JA, Jongejan JA. Quinoproteins, enzymes with pyrrolo-quinoline quinone as cofactor. Annu Rev Biochem. 1989;58:403–26.
Kobe B, Kajava AV. The leucine-rich repeat as a protein recognition motif. Curr Opin Struc Biol. 2001;11:725–32.
Jing H, Takagi J, Liu JH, Lindgren S, Zhang RG, Joachimiak A, et al. Archaeal surface layer proteins contain beta propeller, PKD, and beta helix domains and are related to metazoan cell surface proteins. Structure. 2002;10:1453–64.
Jenkins J, Mayans O, Pickersgill R. Structure and evolution of parallel beta-helix proteins. J Struct Biol. 1998;122:236–46.
Takamiya K, Nishimura M. Dual roles of ubiquinone as primary and secondary-electron acceptors in light-induced electron-transfer in chromatophores of chromatiun-D. Plant Cell Physiol. 1975;16:1061–72.
Ding HG, Robertson DE, Daldal F, Dutton PL. Cytochrome-Bc1 complex [2fe-2s] cluster and its interaction with ubiquinone and ubihydroquinone at the Q0 site - a double-occupancy Q0 site model. Biochemistry. 1992;31:3144–58.
Kato C, Kawai E, Shimizu N, Mikekado T, Kimura F, Miyazawa T, et al. Determination of pyrroloquinoline quinone by enzymatic and LC-MS/MS methods to clarify its levels in foods. Plos One. 2018;13:e0209700.
Noji N, Nakamura T, Kitahata N, Taguchi K, Kudo T, Yoshida S, et al. Simple and sensitive method for pyrroloquinoline quinone (PQQ) analysis in various foods using liquid chromatography/electrospray-lonization tandem mass spectrometry. J Agr Food Chem. 2007;55:7258–63.
Yan Z, Joshi P, Gorski CA, Ferry JG. A biochemical framework for anaerobic oxidation of methane driven by Fe(III)-dependent respiration. Nat Commun. 2018;9:1642.
Gottschalk G, Thauer RK. The Na+-translocating methyltransferase complex from methanogenic archaea. BBA Bioenerg. 2001;1505:28–36.
Abken HJ, Tietze M, Brodersen J, Baumer S, Beifuss U, Deppenmeier U. Isolation and characterization of methanophenazine and function of phenazines in membrane-bound electron transport of Methanosarcina mazei Gol. J Bacteriol. 1998;180:2027–32.
Yee MO, Rotaru AE. Extracellular electron uptake in Methanosarcinales is independent of multiheme c-type cytochromes. Sci Rep. 2020;10:372.
Matsutani M, Yakushi T. Pyrroloquinoline quinone-dependent dehydrogenases of acetic acid bacteria. Appl Microbiol Biot. 2018;102:9531–40.
Qin ZJ, Yu SQ, Chen J, Zhou JW. Dehydrogenases of acetic acid bacteria. Biotechnol Adv. 2022;54:107863.
Anthony C. Pyrroloquinoline quinone (PQQ) and quinoprotein enzymes. Antioxid Redox Sign. 2001;3:757–74.
Holmes DE, Rotaru AE, Ueki T, Shrestha PM, Ferry JG, Lovley DR. Electron and proton flux for carbon dioxide reduction in methanosarcina barkeri during direct interspecies electron transfer. Front Microbiol. 2018;9:03109.
Huang LY, Liu X, Zhang ZS, Ye J, Rensing C, Zhou SG, et al. Light-driven carbon dioxide reduction to methane by Methanosarcina barkeri in an electric syntrophic coculture. ISME J. 2022;16:370–7.
Evans PN, Boyd JA, Leu AO, Woodcroft B, Parks DH, Hugenholtz P, et al. An evolving view of methane metabolism in the Archaea. Nat Rev Microbiol. 2019;17:219–32.
Sowers KR, Baron SF, Ferry JG. Methanosarcina-acetivorans Sp-Nov, an acetotrophic methane-producing bacterium isolated from marine-sediments. Appl Environ Micro. 1984;47:971–8.
Tang J, Zhuang L, Ma JL, Tang ZY, Yu Z, Zhou SG. Secondary mineralization of ferrihydrite affects microbial methanogenesis in geobacter-methanosarcina cocultures. Appl Environ Micro. 2016;82:5869–77.
Li J, Hao XD, van Loosdrecht MCM, Luo YQ, Cao DQ. Effect of humic acids on batch anaerobic digestion of excess sludge. Water Res. 2019;155:431–43.
Song YX, Duan LF, Du KF, Song C, Zhao S, Yuan XZ, et al. Nano zero-valent iron harms methanogenic archaea by interfering with energy conservation and methanogenesis. Environ Sci Nano. 2021;8:3643–54.
Acknowledgements
The authors thank Sen Wang from State Key laboratory of Microbial Technology of Shandong University for help and guidance in SEM. The authors thank Wenshan Qi from Biozeron Co., Ltd. for his technical assistance of RNA-Seq in this study. The authors thank Dr. Lovley’s group for providing ∆hpt and ∆hpt∆mmcA of M. acetivorans. This work was supported by National Natural Science Foundation of China (22008142), Natural Science Foundation of Shandong Province (ZR2022YQ31), Natural Science Foundation of Jiangsu Province (BK20200232), and Qilu Youth Talent Program of Shandong University. Prof. Shuguang Wang acknowledges Taishan Scholars project of Shandong Province (NO. tstp20230604).
Author information
Authors and Affiliations
Contributions
ZY and JF conceived and designed the study; YS, KD, LL, RH and FZ performed the experiment and collected the data; ZY, RH, YS, LL and MW wrote the manuscript; ZY, RH, CS, XY, SZ, MW, SW and JF analyzed and interpreted the data; ZY, RH, CS, XY, SZ, MW, SW and JF revised the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Song, Y., Huang, R., Li, L. et al. Humic acid-dependent respiratory growth of Methanosarcina acetivorans involves pyrroloquinoline quinone. ISME J 17, 2103–2111 (2023). https://doi.org/10.1038/s41396-023-01520-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41396-023-01520-y