Abstract
Average sea surface temperatures are expected to rise 4° this century, and marine phytoplankton and bacterial community composition, biogeochemical rates, and trophic interactions are all expected to change in a future warmer ocean. Thermal experiments typically use constant temperatures; however, weather and hydrography cause marine temperatures to fluctuate on diel cycles and over multiple days. We incubated natural communities of phytoplankton collected from California coastal waters during spring, summer, and fall under present-day and future mean temperatures, using thermal treatments that were either constant or fluctuated on a 48 h cycle. As assayed by marker-gene sequencing, the emergent microbial communities were consistent within each season, except when culture temperatures exceeded the highest temperature recorded in a 10-year local thermal dataset. When temperature treatments exceeded the 10-year maximum the phytoplankton community shifted, becoming dominated by diatom amplicon sequence variants (ASVs) not seen at lower temperatures. When mean temperatures were above the 10-year maximum, constant and fluctuating regimes each selected for different ASVs. These findings suggest coastal microbial communities are largely adapted to the current range of temperatures they experience. They also suggest a general hypothesis whereby multiyear upper temperature limits may represent thresholds, beyond which large community restructurings may occur. Now inevitable future temperature increases that exceed these environmental thresholds, even temporarily, may fundamentally reshape marine microbial communities and therefore the biogeochemical cycles that they mediate.
Similar content being viewed by others
Introduction
Marine phytoplankton draw down atmospheric CO2, support marine food webs, and provide long-term carbon storage in underlying deep waters [1]. Currently, anthropogenic CO2 inputs are inducing changes in phytoplankton communities by changing temperature and climate regimes [2]. Present-day atmospheric CO2 concentrations of >400 ppm have not been seen for almost 12 million years, and these past elevated CO2 events were accompanied by substantial warming [3, 4]. The high specific heat of seawater has meant that warming of the ocean’s surface has happened more slowly than warming of land. Even so, global mean sea surface temperature (SST) has gone up 0.7 °C in the last three decades, and is expected to rise an additional 4 °C this century [3, 5, 6].
Warming influences phytoplankton growth and physiology [7, 8]. Growth rates across temperatures for phytoplankton typically have thermal performance curves (TPC) that increase gradually with rising temperatures to an optimal maximum, then decrease rapidly at higher temperatures [9]. Thermal optima and limits for species are typically connected to in situ thermal regimes. For instance, phytoplankton at lower latitudes often live near their optimal temperatures, whereas temperate species are temperature-limited [10,11,12]. Because of the rapid decrease in growth rates beyond the thermal optimum, rising temperatures can lead to changes in phytoplankton phenology [13, 14], standing photosynthetic biomass [15], metabolic rates, and stoichiometry [16], affecting phytoplankton-mediated biogeochemistry and higher trophic levels. For example, some studies predict diatoms will become less abundant, with cyanobacteria or pico- and nanoeukaryotic taxa being favored [17,18,19], while others suggest the opposite [20, 21]. Shifts within these taxonomic groups are also likely, with warmer conditions selecting for more high-temperature-adapted species or strains [12, 22,23,24].
Typically, studies assessing the impacts of warming on phytoplankton examine shifts in physiological and biogeochemical responses under constant-temperature conditions in a laboratory incubator. In reality, the sea surface is a dynamic thermal environment with temperatures fluctuating over varying time scales from changing weather, diel cycles, vertical and horizontal advection, seasonal changes, and ocean-atmosphere oscillations [25, 26]. Moreover, rising mean atmospheric temperatures are predicted to increase both the frequency and magnitude of future thermal fluctuations in the surface ocean [27, 28]. Because TPCs of phytoplankton and other marine organisms decline quickly past their optimal temperature, thermal fluctuations at higher temperatures may mean periodically experiencing temperatures that are detrimental to growth [29].
Despite the growing recognition that temperature variations and transitory extreme thermal events need to be considered when making predictions about biological responses to climate change, few studies have looked at how fluctuating temperatures impact microbial communities. One of these studies found that growth rates could either increase or decrease under fluctuating temperatures, depending where they occurred on the phytoplankton’s TPC [30]. Communities in freshwater lakes have shown shifts towards smaller, faster-growing taxa [31]. Variable temperature regimes also can help maintain crucial diversity by supporting multiple thermal niches [32]. Long-term evolution of the cosmopolitan marine phytoplankton Ostreococcus tauri under fluctuating pCO2 favored greater phenotypic plasticity, which was then found to be predictive of future evolutionary success [33]. Similar results were seen in diatoms maintained for >100 generations under fluctuating temperatures [34]. Nutrient availability can also interact with environmental variability. Fluctuating temperatures decrease growth of the nitrogen-fixing cyanobacterium Trichodesmium under nutrient-replete conditions, but growth rates are lower and similar in variable and constant thermal treatments under phosphorus limitation [35].
We examined how seasonally changing California coastal plankton microbial communities responded to fluctuating temperatures over 7–14 generations in a series of incubation experiments. These included both constant and fluctuating temperatures, at both present-day and projected-future means. Phytoplankton assemblages in coastal California experience both seasonal and short-term temperature fluctuations, with daily fluctuations of up to 4 °C being common [25, 36, 37], suggesting they may be well-adapted to thermal variability. Additionally, microbial communities at our coastal sample collection site (San Pedro Ocean Time-series; SPOT) have well documented seasonal and annually recurring patterns [37,38,39], allowing us to interpret our temperature manipulations in the context of 20 years of microbial community and environmental data.
Methods
Sampling site
Surface water for the experiment was collected from the Southern California Bight at the San Pedro Ocean Timeseries (SPOT) station (33˚33′ N, 118˚24′ W). Seasonal sampling in September 2016 (summer), November 2016 (fall), and May 2017 (spring) examined microbial communities collected at ambient surface water temperatures of 20.6°, 16.5° and 16.1°, respectively (Table 1). Seawater was collected in carboys from 3 m depth, with 100 µm mesh prefiltration to remove zooplankton, and was then taken back to the University of Southern California, where it was stored overnight at collection temperature. Initial samples and the incubation-experiment setup used water combined from all the collection carboys. The remaining surface water was filtered through a 0.2 µm gravity filter and used for subsequent culture dilutions.
Incubation experiments
Because of the oligotrophic conditions and low biomass at SPOT, nutrients were added to stimulate photoautotrophic growth and so enable measurements of the effects of temperature on microbial communities. Each incubation experiment used triplicate one-liter flasks enriched with nitrate, silicate and phosphate added to final concentrations of 30, 30 and 2 µM, respectively, representing values often observed during strong upwelling events along the California coast [40]. Iron, other trace metals, and vitamins were added at replete concentrations equivalent to Aquil medium [41] to avoid micronutrient limitation.
We measured the overall temperature responses of each seasonal phytoplankton community by generating thermal performance curves (TPCs). Initial seawater was split into 8−10 temperature treatments, ranging from 10 to 32 °C. Individual water baths with their own heating elements and thermostats were used to ensure accurate temperature treatments. Due to these being mixed communities, in vivo Chl a fluorescence was used to calculate bulk phytoplankton community growth rates for all temperatures using the formula: \(\mu = \frac{{\ln \left( {F_1 \ \div \ F_0} \right)}}{{t_1 \ - \ t_0}}\). In this equation, µ is divisions per day, F0 and F1 are the raw fluorescence unit values measured on a Turner AU-10 fluorometer (Turner Designs Inc., Sunnyvale, CA, USA) at the beginning and end of each dilution period respectively, and t is the number of days between measurements. We fitted the TPC growth data to the Eppley−Norberg temperature model to make predictions of each community’s thermal limits and optimal growth temperature [10]. We also used a modified version of this model that takes into account thermal fluctuations to estimate growth rates in variable thermal environments [42] to compare our experimental variable thermal treatments (±4 °C) to 10 years of daily maximum SST data obtained from the National Ocean and Atmospheric Administration’s National Data Buoy Center using a mooring station located 10.8 km from SPOT.
Additionally, we split enriched SPOT water into temperature treatments intended to simulate present and predicted-future surface-water temperatures at both constant and fluctuating temperatures (Table 1). Present temperatures were set to match the temperature at SPOT at the time of collection. The present-constant treatments acted as our experimental control. Future temperatures were increased 4 °C (spring and fall) or 5 °C (summer) from the present temperature (Table 1), following predicted increases in SST [6]. The two fluctuating temperature treatments had the same means as the constant treatments, but they alternated between a warm phase above (+4 °C) and a cold phase below (−4 °C) the mean value sequentially every 24 h, yielding a 48 h complete thermal cycle (Fig. S1). This time scale was based on previously published high-resolution temperature data from the Southern California Bight [25]. Hereafter we will refer to pre-nutrient addition samples as “initial” and each seasonal temperature study as “experimental”.
Bulk growth was monitored using daily in vivo fluorescence measurements, and one-part culture was transferred to ten-parts filtered, enriched seawater every 2 days to keep cultures in logarithmic growth and maintain a relatively constant nutrient environment [10, 43]. All experiments were run for at least seven transfers (~7−14 doublings over at least 14 days, depending on growth rate). After seven dilutions, experiments were sampled daily for 3 days to look for responses specific to either cold or warm phase of fluctuation treatments (Fig. S1). Due to high growth rates during the summer experiment, an additional dilution had to be made between cold and warm fluctuations to avoid cells entering stationary phase. For experiments we calculated growth rates using the same equation given above, but in addition to in vivo Chl a fluorescence, growth rates were also calculated based on particulate organic carbon (POC), intended to represent the whole assemblage, and biogenic silica (BSi) as a metric for diatom-specific rates.
Biogeochemical assays
To measure POC and particulate organic nitrogen (PON), samples were filtered onto precombusted GF/F filters (2 h at 450 °C) and analyzed using a Costech Elemental Combustion system (Valencia, CA, USA) [44]. POC was used to estimate bulk assemblage growth rates by recording the change in POC over 2 days, capturing growth during both cool and warm periods. To measure BSi, samples were filtered onto 3 µm polycarbonate filters and measured as in [45] to estimate diatom biomass. Similar to POC-derived growth rates, changes in BSi over 2 days were used to derive diatom-specific growth rates in our treatments. Cells were filtered onto precombusted GF/F filters for particulate organic phosphorus (POP) measurements [46]. In addition to indirectly measuring Chl a using in vivo fluorescence, we measured total chlorophyll by filtering onto GF/F filters and extracting in 90% acetone for 24 h. Extracted Chl a and in vivo fluorescence were measured on a Turner AU-10 (Turner Designs Inc., Sunnyvale, CA). During spring and fall experiments, carbon fixation rates were measured by spiking 30 ml of each enrichment with 50 µl of 14C labeled sodium bicarbonate, then incubated for 3 h, filtered onto GF/F filters, and placed in 4.5 ml of scintillation solution. Total radioactivity (TA) was measured using triplicate solutions of combined isotope and scintillation solution spiked with 100 µl of phenyethlamine. We accounted for filter absorption using 10 ml from each replicate-enrichment spiked with identical amounts of isotopes and filtered immediately. Samples were incubated in the dark overnight and radioactivity was measured with a Tri-Carb 2500TR liquid scintillation counter after 24 h (Beckman Coulter Inc., Brea, CA) [47].
Amplicon sequencing
Microbial diversity was sampled before nutrients were added, and at the end of the final temperature fluctuation cycle. Cells were filtered (1.2 µm polycarbonate) and stored in liquid nitrogen. Extractions used the DNeasy Power Soil kit (Qiagen, Hilden, Germany) modified to include a 10-min 65 °C incubation before vortexing. Amplification and sequencing of the V4-V5 hypervariable region of the 16S rRNA gene was done using the primers 515F-Y (5′-GTGYCAGCMGCCGCGGTAA-3′) and 926R (5′-CCGYCAATTYMTTTRAGTTT-3′), as described in [48]. These primers successfully amplify large proportions of known prokaryotes, and chloroplasts (via the 16S rRNA gene), as well as eukaryotes (via the 18S rRNA) [48]. These primers have been previously used to describe microbial communities at this study site [49]. Library prep and sequencing was done at Molecular Research DNA labs (MR DNA; Shallowater, TX, USA) on the Illumina Miseq platform producing 2 × 300 bp paired-end reads. DNA samples from the spring experiment were treated the same way as summer and fall DNA samples, but were sequenced on a different date. To avoid potential sequencing run-specific batch effects, each season was analyzed individually. The quality of DNA from one replicate in the spring future-constant treatment was low and consequently contained few reads. This replicate was excluded from sequence analysis.
Sequence data analysis
Our processing workflow is shown in Fig. S2. In short, raw sequence reads were demultiplexed using Sabre (github.com/najoshi/sabre, version 1.0) and primers removed with usearch’s fastx_truncate (version 9.2) command to cut the first 20 bases from forward and first 19 from reverse reads. These were analyzed with the DADA2 pipeline, version 1.6.0 [50]. Default settings were used except where noted following DADA2’s standard workflow, version 1.8 (https://benjjneb.github.io/dada2/tutorial.html). The 18S rRNA gene region that is amplified by these primers is typically longer than our 2 × 300 paired-end sequencing protocol spans, and hence would be discarded by the merge step our analysis pipeline [48]. We gathered all nonmerged reads and separated out putative 18S rRNA gene sequences from reads rejected due to quality issues using the number of mismatched base pairs. We found that sequences with >35 mismatches typically matched eukaryotic 18S rRNA gene sequences when BLASTed against NCBI’s nonredundant nucleotide (nr/nt) database (Table S1). Because of this correlation, all reads with >35 mismatches were assumed to be eukaryotic 18S rRNA gene sequences. These were concatenated with ten ambiguous, “n” bases and processed with the merged, 16S rRNA reads. Amplicon sequence variants (ASVs) identified by the Silva132 database [51] as chloroplasts were removed and separately assigned taxonomy using PhytoREF, a curated database of phytoplankton chloroplast 16S rRNA sequences [52]. ASVs derived from putative 18S rRNA gene sequences were identified using the Protist Ribosomal Reference database, PR2 [53]. Previous work with these primers at SPOT observed that estimates of phytoplankton diversity were typically similar using reads assigned to either 16S or 18S rRNA amplicons [49]. For this study we chose to rely on the 16S rRNA plastid gene sequences to describe the eukaryotic phytoplankton community. 18S rRNA gene copy number varies considerably between taxa based on genome size [54], whereas plastid numbers can vary based on environmental variables (e.g. nutrient availability) which we control for in our experiments. Putative-18S assigned taxonomy was used to check for the presence of metazoan grazers (which should have been largely removed with our prefiltering during sampling), and for dinoflagellates, whose plastid 16S rRNA gene sequences are highly divergent [55] and not amplified with these primers. For the highest level of taxonomic resolution, we BLASTed all bacterial ASVs that comprised >10% and eukaryotic ASVs comprising >5% of recovered reads in any given sample against the NCBI’s nr/nt database, excluding uncultured sample sequences. We used percent similarity thresholds to assign taxonomic rank following published values for primers amplifying the V4 rRNA region [56]. For dominant phytoplankton ASVs we confirmed their taxonomic identity by also BLASTing the corresponding 18S rRNA sequences. Because these primers produce relatively fewer 18S rRNA amplicons and the natural differences in copy numbers of each gene, we paired sequences by comparing the relative abundance of recovered 18S and plastid 16S rRNA sequences. ASVs of each gene that had the strongest correlation (highest Spearman’s correlational coefficient) were considered to belong to the same organism.
Analysis of read counts including calculations of diversity, ecological distance, and all ordinations was done in R [57] and RStudio [58] using the Phyloseq [59] and Vegan [60] packages. For ordinations, the ASV count matrix was first transformed using the variance stabilizing transformation within DESeq2 [61], Euclidean distances were calculated (Vegan), and we tested for significance between groups using a permutational ANOVA (Vegan), after confirming an equal amount of variance between groups using the betadisper function (Vegan). Significance of temperature treatment effects on major taxonomic groups was tested using both one-way ANOVA and t test, and DESeq2 was used to test for differential abundance in individual ASVs by comparing them to our control treatment.
Results
During each sampling effort, conditions at SPOT were oligotrophic. Total chlorophyll levels were <0.5 µg/l, with little available nitrogen (mean = 0.026 µM, SD = 0.01) or phosphorus (mean = 0.17 µM, SD = 0.03) (Table S2). Thermal response curves showed the seasonal assemblages in all three experiments were able to grow at least 6 °C above the 10-year record maximum temperature for each season at SPOT (Fig. 1). Bulk growth rates of the spring communities did not drop to zero until 32 °C was reached (Fig. 1a), and growth rates in the summer (Fig. 1b) and fall (Fig. 1c) were still maximal at the highest temperature tested (30 °C). Maximum growth rates for each seasonal community were measured at or above 26 °C (Fig. 1).
Chlorophyll a fluorescence-based thermal performance curves calculated for the seasonal mixed phytoplankton community at SPOT during a spring (n = 13, R2 = 0.74), b summer (n = 8, R2 = 0.77), c fall (n = 10, R2 = 0.76). Fitted line shows the Eppley−Norberg temperature model. Vertical lines summarize 10 years of seasonal temperature data, where the dark solid line is the mean with dashed lines ± one standard deviation, and the solid red line is the highest recorded temperature
For each experiment, we compared the impact of our ±4 °C experimental fluctuations to daily temperature fluctuations in our 10-year temperature dataset from SPOT (Table S3). The modified Eppley−Norberg model of Bernhardt et al. that accounts for temperature fluctuations [42] predicted no difference between in situ temperature fluctuations and our ±4 °C fluctuations. This suggests that from the perspective of phytoplankton growth rates, our temperature fluctuations were similar to what they experience in situ.
In the spring experiment, growth rates calculated using POC were lower in the present-fluctuating (12–20 °C) than the present-constant (Fig. 2a, p = 0.03), but this difference was not reflected in either the BSi-derived diatom growth rate (Fig. 2d) or the BSi:POC ratio (Fig. 2g). Future-fluctuating temperatures in the summer (22–30 °C) had POC-derived growth rates (Fig. 2b) significantly higher than the present-constant (21 °C, p = 0.004) and present-fluctuating treatment (17–25 °C, p = 0.05). In the summer, BSi-derived growth rates were higher in the future-constant treatment than other treatments (p < 0.05). In the fall, treatment had no effect on POC or BSi-derived growth rate (Fig. 2c, f), but the future-constant treatment (20 °C) had a significantly higher BSi:POC ratio than the other treatments (Fig. 2i, p < 0.05). Other biogeochemical and bulk biochemical parameters were relatively unaffected by the temperature manipulation experiments. Elemental ratios of particulate C, N, P, Chl a content, and carbon fixation rates were variable, but not statistically different for treatments within each seasonal experiment (Fig. S3).
Community growth rates determined as changes in particulate organic carbon (POC, a–c), and the diatom-specific growth measured with changing BSI (d–f), both measured over 2 days at the end of 14 days of growth. g−i depict BSi to POC ratios as an indicator of the relative abundance of diatoms in each treatment. Statistical significance between two treatments (p < 0.05) is shown with a star. p values for nearly significant treatments (p < 0.1) are shown with their respective brackets, and error bars represent standard deviation
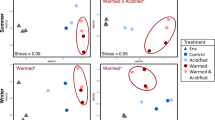
rRNA-gene amplicon sequencing was used to assess changes in the phytoplankton communities associated with different temperature treatments. Ordinations clearly separated initial and experimental samples, based on nutrient level (Fig. S4A). When initial samples were removed, collection-season clearly correlated with sample groupings (Fig. S4B). Within each experiment, the first two axes of a principle coordinates analysis (PCoA) were able to explain between 27.7% and 32.4% of the variation, and each seasonal study had a significant difference between at least two treatments as detected by a permutational ANOVA (Fig. 3). In all treatments, bacterial sequences made up the majority of recovered amplicons (Fig. 4a). Chloroplast 16S rRNA gene sequences belonged mostly to picoeukaryotes in initial samples, with diatoms being dominant in spring and fall experiments (Fig. 4a, b).
a Bars comparing the percentage of recovered rRNA sequences, largely representing heterotrophic bacteria and two major functional groups of autotrophic phytoplankton. Values are expressed as the mean percent amplicons recruited for bacteria (black), diatoms (gray), and picoeukaryotic phytoplankton (light gray). Error bars represent standard deviation. b Phylum and class level (proteobacteria) abundance using 16S rRNA gene sequence identity. The size of each bubble corresponds to percent amplicons recovered. Closed circles show taxa that are significantly more or less abundant by a one-way ANOVA (p < 0.05). Brackets show specific significant differences between groups. Plastid 16S rRNA gene sequences were used for taxonomic assignments of eukaryotic phytoplankton
The majority of recovered sequences in all experiments belonged to the Alpha and Gammaproteobacteria and the Bacteroidetes phyla (Fig. 4b). Within these larger taxonomic groups, 132 bacterial ASVs were observed that comprised >1% of the relative abundance of amplicons recovered from at least one sample (Table S4). The majority of these belonged to the Bacteroidetes (61 ASVs) with Saprospiraceae (22 ASVs) and the Flavobacteriaceae (18 ASVs) being the most diverse families. Alphaproteobacteria included 44 ASVs, which were largely made up by the Rhodobacteraceae (21 ASVs). Of the 15 Gammaproteobacteria comprising >1%, ten were in the genus Alteromonas. Three ASVs were placed into Gammaproteobacteria; however, SILVA was only able to identify two of them to class. The other ASV was placed into the family Oligoflexaceae. Other groups crossing the 1% threshold were the Planctomycetes (three ASVs), Verrucomictrobia (two ASVs), and the Actinobacteria (one ASV).
We did not detect differences in community composition between constant vs. fluctuating treatments at broad taxonomic levels in the spring experiment (Fig. 4b), but did for Alphaproteobacteria and Cyanobacteria in the fall (p < 0.05). The summer experiment demonstrated much greater variation between the constant temperature and fluctuating treatments. For example, summer-derived communities demonstrated a decrease in the relative abundance of Alphaproteobacteria recovered sequences (p < 0.05) in the future-fluctuating treatment as compared to both present-constant and present-fluctuating treatments. These were mostly made up of copiotrophic heterotrophs in the Hyphomonadaceae and Rhodobacteraceae (Table S4), and there was also an increase in the relative abundance of Planctomycetes (p < 0.05) sequences in the future-fluctuating treatment. The shift in the Planctomycetes was from an increase in a single ASV (ASV20), which when BLASTed most closely matched Candidatus Brocadiales fulgida, a known anammox bacteria [62]. Gammaproteobacteria also decreased in relative abundance (p < 0.05) during the summer experiment in the present-fluctuating treatment relative to the present-day constant treatment. The largest change came from a single ASV (asv3) identified with BLAST as an Alteromonas sp. (Fig. S5). In the spring, two bacterial ASVs that made up more than 10% of the recovered bacterial amplicons belonging to the genus Phaeobacter and an unknown Flavobacteriaceae declined in relative abundance in all treatments, compared to the present-constant control (Fig. S5A). Similarly, in the summer, any temperature manipulation caused two dominant bacterial ASVs (one Alteromonas sp. and another unknown Flavobacteriaceae) to decrease their relative abundance.
Bacillariophyta were the most abundant eukaryotic phytoplankton in experiments, although some prymnesiophytes were detected (Fig. 4b). 18S sequences were screened for dinoflagellates and for the presence of any metazoan grazers not removed by prefiltering (Fig. S6). Neither was seen except in initial samples, particularly in the spring experiments, where copepods (likely nauplii) made up as much as 35% of total initial sequences recovered (Fig. S6). In the summer experiment, prymnesiophyceae sequences were most abundantly recovered in the present-constant (2.1%, SD = 0.7) compared to other treatments.
The phytoplankton community in each incubation experiment had one or two dominant ASVs making up the majority of the photoautotrophic community (Fig. 5, Table S5). In the spring experiment an uncharacterized Pseudo-nitzschia sp. (asv9) and Minidiscus trioculatus (a small, unicellular, centric diatom; asv2) were the most abundant in all samples (Fig. 5). The Pseudo-nitzschia 18S sequence recovered (asv105; Fig. S7A; Table S6) was 100% identical to Pseudo-nitzschia americana strain UNC1412 (NCBI accession number KX229689.1), which was isolated from the California upwelling zone in 2014. On a phylogenetic tree of top BLAST hits, the two nearest references sequences were not known producers of the toxin domoic acid (Fig. S8), which is produced by many other members of this genus. The same Minidiscus trioculatus ASV (asv2) was the most abundant phytoplankton in each of the fall enrichments (Fig. 5b, Fig. S7B).
Heatmaps of amplicon sequence variants (ASVs) that comprised: a >10% of total reads within a sample for bacteria, and >5% of the total reads for b diatoms and c picoeukaryotic phytoplankton. Triangles represent treatments where a given ASV was differentially abundant compared to the control (present-constant treatment outlined with dashed lines)
In the summer experiment, both present-constant and present-fluctuating treatments were largely dominated by an ASV matching the picoeukaryotic phytoplankton (asv1) belonging to the group basal to the diatoms, the bolidophytes; however, the recovered 18S rRNA amplicon was a 100% match to the diatom Leptocylindrus convexus, part of an early branching group of diatoms [63, 64]. Microscopy revealed large numbers of cells morphologically identical to Leptocylindrus, so asv1 was assigned taxonomy based on its 18S rRNA gene. In the warmer, future-constant treatment this diatom is supplanted by a different diatom ASV Chaetoceros simplex (asv8), which was in turn replaced by another diatom, Arcocellulus mammifer (asv4), when those warmer temperatures fluctuated (Figs. 5b, S5). The matching 18S rRNA sequence recovered for asv4 belonged to the genus Minutocellus (Fig. S7C, Table S6). Because Arcocellulus and Minutocellus are very closely related [63], we used the plastid 16S rRNA sequence identity to assign its taxonomy.
Testing of differential abundance using DESeq2 [61] compared all treatments to the present-day, constant control treatment. Some statistically significant shifts in abundance were measured in every experiment (Table S7); however, in general most of these shifts between treatments did not involve dominant ASVs (>10% for bacteria and >5% for eukaryotic phytoplankton). Those that did were the increase in ASVs matching the genus Rugeria (asv31) in the spring future-fluctuating treatment (Fig. 5a), and the shift from a Leptosylindrus-dominated to a Chaetoceros-dominated (future-constant) and Arcocellulus-dominated (future-fluctuating) phytoplankton community in the summer (Fig. 5b).
Discussion
Enrichments showed positive growth rates even at temperatures higher than those ever experienced at the coastal California site where they were collected (>25°). In addition, the bulk community growth rates were relatively stable across temperatures and did not show the typical negative skewness of phytoplankton isolate thermal response curves [11]. This broad optimum range across temperatures is likely the result of thermal functional redundancy in these natural communities. In other words, different members likely have different limits defining their optimum and stressful temperatures, thus providing redundancy and robustness to bulk growth rates, even with a shifting underlying community. Careful examination of the communities in our comparison of warming and fluctuating conditions showed that incubations largely maintained the same dominant taxa until they were exposed transiently to unusually high temperatures exceeding the summer experiment upper temperature limit.
In our experiments, no consistent difference was observed in our POC- and BSi-determined growth rates between constant and fluctuating treatments. Thermal fluctuations and nutrient inputs are often linked through vertical mixing and advection events in the coastal regime where SPOT is located, so the phytoplankton we enriched for may already be well-adapted to fluctuating temperatures. At future mean temperatures in both the summer and fall (26 and 20 °C respectively), we did observe significant and near significant differences between constant and fluctuating conditions in the ratio of diatom frustule mass (BSi) relative to the POC present. Plotting these ratios during the final dilution series (3 days) shows that BSi accumulated faster relative to POC in the future-constant treatment in the summer and fall experiments, than in future-fluctuating treatments (Fig. S9B, C). This difference was particularly pronounced in the summer experiment, when the slope of a line fitted to the data was 6.5 times higher in the future-constant vs. the future-fluctuating treatment (Fig. S9B).
Our experimental design was intended to simulate climate change impacts on primary producers within a coastal zone. Outside of a brief spring bloom, SPOT is typically oligotrophic, with new primary production often relying on episodic and ephemeral Ekman upwelling [36]. The low chlorophyll, picoeukaryote-dominated initial community shifted to a high chlorophyll, largely diatom-dominated community in our experiment, which is what we would expect following a typical transitory upwelling or mixing event at the SPOT time series station. Because of the strong influence of the experimental nutrient additions, initial communities were excluded from any statistical testing.
The communities we observed post nutrient addition were distinct to each season. The differences in the outcomes of fall and spring incubations, despite similar temperatures, are likely due to differences in the communities when the samples were collected. The microbial community at SPOT when sampled monthly over multiple years is most dissimilar to the microbial community 6 months before or after the sample is taken [38]. Our fall and spring samples were taken exactly 6 months apart, so it is not surprising that after the incubations we got different results. Of course, as also suggested by our results, these distinct spring and fall community structures may be largely a function of their differing recent thermal histories.
Bacterial communities in all three experiments were consistent with those often associated with diatom blooms. For instance, ASVs from genera such as Pseudophaeobacter (asv16), Phaeobacter (asv18), and Rugeria (asv31, Alphaproteobacteria) as well as Alteromonas (asv3, Gammaproteobacteria) and Lewinella (asv32 and asv26, Bacteroidetes) all made up 10% or more of the amplicons in any one sample. These genera are copiotrophic heterotrophs, and so are frequently reported in diatom cultures and in situ blooms [65]. Interestingly, a Bacteroidetes ASV matching Kordia jejudonensis (asv11) was only relatively abundant in the spring present-fluctuating treatment, in which diatom levels were the lowest within that experiment. This genus contains species that produce allelopathic compounds known to be lethal to phytoplankton, which could explain the relatively low diatom counts [66].
Phytoplankton species that emerged following nutrient additions also were typical of eutrophic periods in the California Current. Pseudo-nitzschia, Minidiscus, Leptocylindrus, and Chaetoceros are common bloom-forming diatom genera in the California Current System [67, 68]. The dominant diatom from the future-fluctuating treatment in the summer, Arcocellulus mammifer, is not mentioned in the literature at this site. Instead, blooms of this species have been recorded in aquaculture ponds in the tropical South Pacific [69]. The apparent thermophilic niche of this species is consistent with the fact that it was only observed in our treatment intermittently exposed to extreme high temperatures.
Leptocylindrus convexus sequences were only recovered in relative abundance at present-constant and present-future temperatures. In the future-constant treatment from the same experiment (mean temperature = 26 °C), this organism seemed to be supplanted by the chain forming diatom Chaetoceros simplex. Chaetoceros spp. are heavily silicified, potentially explaining the significantly higher BSi:C ratio in the future-constant treatment. This species was only dominant in the future-constant treatment, however, and in the future-fluctuating treatment the dominant phytoplankter changed again to the diatom Arcocellulus mammifer. This shift was also seen in declining diatom-specific growth rates and BSi concentrations relative to the future-constant treatment.
In our experiment the phytoplankton bloom from each seasonal enrichment was distinct from those collected during the other seasons. Past work at SPOT has showed that microbial assemblages from a given month are most similar to other months from the same season, even across years (378). Further, we compared the abundance of the dominant phytoplankton taxa from our spring (ASV9 Pseudo-nitzschia sp.) and summer (asv1 Leptocylindrus convexus, asv8 Chaetoceros simplex, and asv4 Arcocellulus mammifer) incubation experiments with previously published data from Needham and Fuhrman (2016; 49), who followed the response to a spring-time nutrient pulse over the course of 6 months (Fig. S10). All three dominant ASVs from our spring incubations were detected in nearly every in situ sample from this prior study. In March when temperatures were low and available nitrogen high (>4 µM), amplicons matching ASV9 (>99% similarity across the entire length of the sequence) became more abundant, while amplicons matching ASV1 or ASV8 remained barely detectable. In mid-May in the Needham and Fuhrman (2016; 49) study as temperatures began to rise, a modest increase in available nitrogen (~1 µM) stimulated chlorophyll a production and resulted in a small increase in amplicons matching asv1 and asv8 that were dominant in our summer experiment, while those matching ASV9 remained low. In addition, asv4 that was dominant in the summer future-fluctuating treatment, where it experienced much higher temperatures than those observed at SPOT, remained the same throughout this dataset. The consistency between our experimental community structure and in situ observations suggests that it is the seasonal thermal environment that dictates which species are able to respond to ephemeral nutrient inputs, and that our experiments accurately simulated these seasonal patterns.
With these experiments, we repeatedly observed that incubation temperatures (whether fluctuating or constant) that fell within present-day norms stimulated dense phytoplankton blooms that were largely taxonomically indistinguishable across all treatments. Similarities in composition also suggest a functional redundancy that maintained biogeochemical and bulk biochemical processes within the envelope of historic temperatures. Even when communities were grown under conditions that periodically exposed them to temperatures close to this maximum (17–25 °C), the dominant primary producers did not change. Only when consistently culturing at high temperatures (26 °C) that exceeded historic maximum temperature (25.3 °C as measured over the past 10 years) did the community of primary producers shift. Further, periodically exposing the community to 30 °C (4.7 °C above the 10-year maximum) pushed the Chaetoceros simplex beyond their thermal maximum, and resulted in enriched abundance of Arcocellulus mammifer regardless of the brevity of exposure and the shift back to lower temperatures (22 °C) the following day.
Thermal impacts such as shifts in dominant taxa or declining phytoplankton abundance could happen at even lower temperatures in situ. For instance, some grazers are less susceptible to thermal stress than eukaryotic phototrophs [70, 71]. Because we removed these from our system, we are unable to assess the interaction of temperature and grazing pressure in shaping community composition. Nutrient levels could also be confounding, as recent work suggests an interactive effect between nutrient concentrations and temperatures, with less thermal resilience under oligotrophic conditions [72], although limiting nutrients can increase thermal tolerance in some marine diazotrophs [73]. Our nutrient concentrations were kept replete through frequent dilutions with nutrient-amended seawater, masking any potential temperature/nutrient availability interactions in our future treatments. It is possible that in situ grazing pressures and nutrient limitation could interact strongly with warming to allow impacts on phytoplankton communities with smaller temperature perturbations.
These data suggest that temperatures that match or slightly exceed historic high temperatures even briefly (on a timescale of weeks) can cause major shifts in dominant phytoplankton, even under nutrient-replete conditions. This is in addition to the impacts of long-term elevated mean temperatures such as those that have been predicted by ocean/atmosphere models [6]. Our observations suggest these new communities are stable, and still capable of maintaining their role in marine ecosystems. This is also consistent with observations from other marine ecosystems, where short-term heat waves have had ecological consequences over and above those of more modest, longer-term warming. A temperature anomaly off Australia’s west coast in 2010/2011 increased temperatures 2.5 °C above seasonal norms, and for a brief period (~1 week) exceeded the normal seasonal temperatures by 5 °C [74]. The effects of these anomalous conditions seemingly irreversibly shifted the ecosystem from a kelp-dominated community with an abundance of temperate fish to a benthic, turf algal assemblage with tropical fish species that were not present before the heatwave. Recent studies that experimentally manipulated thermal regimes with the marine copepod Tigriopus californicus suggested how this process might happen, hypothesizing that prior exposure to sub-lethal warm temperatures made individuals more vulnerable to short-term extreme heat events [75]. This makes sense given the typical shape of microbial thermal curves and the unequal impacts that thermal fluctuations have on growth rates at higher temperature, where they can often decrease optimal and lethal thermal limits.
As marine microbial ecosystems continue to experience warming, it is likely that these scenarios combining warming with fluctuating, short-term heat waves could become more common. This work suggests that encounters with unprecedented high-temperatures could lead to broad shifts in dominant phytoplankton taxa. By simulating the bloom-forming conditions that periodically occur in this coastal regime following upwelling, we observed that the onset of high temperatures not previously experienced in situ may serve to delineate a threshold where warming affects the composition of the microbial community. This threshold is likely modulated by other co-stressors such as nutrient availability and grazing, by the duration of high-temperature exposure, and potentially by the range of temperatures in a given regions of the ocean. Here we offer a testable hypothesis that we believe can act as a starting point for testing the limits of present-day community structure and function in the context of a warming ocean.
Data Availability
All scripts used for quality control, analysis of sequence data, and figure preparation can be found at: https://doi.org/10.6084/m9.figshare.7603790.v2. Sequence data have been uploaded to NCBI under the Bioproject ID PRJNA512541. SRA accession numbers and associated metadata are found in Table S8. Data from San Pedro Ocean Time-series (SPOT) monthly sampling can be found at https://dornsife.usc.edu/spot/data/, and daily temperature data is from the National Data Buoy Center (https://www.ndbc.noaa.gov/), station 46222.
References
Falkowski P, Barber R, Smetacek V. Biogeochemical controls and feedbacks on ocean primary production. Science. 1998;281:200–7.
Hutchins DA, Fu F. Microorganisms and ocean global change. Nat Microbiol. 2017;2:17058.
Hansen J, Sato M, Ruedy R, Lo K, Lea DW, Medina-Elizade M. Global temperature change. Proc Natl Acad Sci USA. 2006;103:14288–93.
Tripati AK, Roberts CD, Eagle RA. Coupling of CO2 and ice sheet stability over major climate transitions of the last 20 million years. Science. 2009;326:1394–7.
Levitus S, Antonov JI, Boyer TP, Locarnini RA, Garcia HE, Mishonov AV. Global ocean heat content 1955–2008 in light of recently revealed instrumentation problems. Geophys Res Lett. 2009;36:1–7.
IPCC. Climate change 2013: The physical science basis. In: Stocker TF, Qin D, Plattner GK, Tignor M, Allen SK, Boschung J, et al., editors. Contribution of Working Group I to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change. Cambridge: Cambridge University Press; 2013.
Eppley RW. Temperature and phytoplankton growth in the sea. Fish Bull. 1972;70:1063–85.
Raven JA, Geider RJ. Temperature and algal growth. New Phytol. 1988;110:441–61.
Norberg J. Biodiversity and ecosystem functioning: a complex adaptive systems approach. Limnol Oceanogr. 2004;49:1269–77.
Thomas MK, Kremer CT, Klausmeier CA, Litchman E. A global pattern of thermal adaptation in marine phytoplankton. Science. 2012;338:1085–8.
Boyd PW, Rynearson TA, Armstrong EA, Fu F, Hayashi K, Hu Z, et al. Marine phytoplankton temperature versus growth responses from polar to tropical waters—outcome of a scientific community-wide study. PLoS ONE. 2013;8:e63091–17.
Fu FX, Yu E, Garcia NS, Gale J, Luo Y, Webb EA, et al. Differing responses of marine N2 fixers to warming and consequences for future diazotroph community structure. Aquat Micro Ecol. 2014;72:33–46.
Gittings JA, Raitsos DE, Krokos G, Hoteit I. Impacts of warming on phytoplankton abundance and phenology in a typical tropical marine ecosystem. Sci Rep. 2018;8:1–12.
Poloczanska ES, Burrows MT, Brown CJ, García Molinos J, Halpern BS, Hoegh-Guldberg O, et al. Responses of marine organisms to climate change across oceans. Front Mar Sci. 2016;3:515–21.
Yvon-Durocher G, Montoya JM, Trimmer M, Woodward G. Warming alters the size spectrum and shifts the distribution of biomass in freshwater ecosystems. Glob Change Biol. 2010;17:1681–94.
Benner I, Diner RE, Lefebvre SC, Li D, Komada T, Carpenter EJ, et al. Emiliania huxleyi increases calcification but not expression of calcification-related genes in long-term exposure to elevated temperature and pCO2. Philos Trans R Soc Lond, B, Biol Sci. 2013;368:20130049–9.
Hare CE, Leblanc K, DiTullio GR, Kudela RM, Zhang Y, Lee PA, et al. Consequences of increased temperature and CO2 for phytoplankton community structure in the Bering Sea. Mar Ecol Prog Ser. 2007;352:9–16.
Feng Y, Hare CE, Leblanc K, Rose JM, Zhang Y, DiTullio GR, et al. Effects of increased pCO2 and temperature on the North Atlantic spring bloom. I. The phytoplankton community and biogeochemical response. Mar Ecol Prog Ser. 2009;388:13–25.
Lewandowska A, Sommer U. Climate change and the spring bloom: a mesocosm study on the influence of light and temperature on phytoplankton and mesozooplankton. Mar Ecol Prog Ser. 2010;405:101–11.
Hinder SL, Hays GC, Edwards M, Roberts EC, Walne AW, Gravenor MB. Changes in marine dinoflagellate and diatom abundance under climate change. Nat Clim Change. 2012;2:271–5.
Zhu Z, Xu K, Fu F, Spackeen JL, Bronk DA, Hutchins DA. A comparative study of iron and temperature interactive effects on diatoms and Phaeocystis antarctica from the Ross Sea, Antarctica. Mar Ecol Prog Ser. 2016;550:39–51.
Kremp A, Godhe A, Egardt J, Dupont S, Suikkanen S, Casabianca S, et al. Intraspecific variability in the response of bloom-forming marine microalgae to changed climate conditions. Ecol Evol. 2012;2:1195–207.
Canesi KL, Rynearson TA. Temporal variation of Skeletonema community composition from a long-term time series in Narragansett Bay identified using high-throughput DNA sequencing. Mar Ecol Prog Ser. 2016;556:1–16.
Demory D, Baudoux A-C, Monier A, Simon N, Six C, Ge P, et al. Picoeukaryotes of the Micromonas genus: sentinels of a warming ocean. ISME J. 2018;305:1–15.
Leinweber A, Gruber N, Frenzel H, Friederich GE, Chavez FP. Diurnal carbon cycling in the surface ocean and lower atmosphere of Santa Monica Bay, California. Geophys Res Lett. 2009;36:L08601–5.
Doblin MA, van Sebille E. Drift in ocean currents impacts intergenerational microbial exposure to temperature. Proc Natl Acad Sci USA. 2016;13:5700–5.
Salinger MJ. Climate variability and change: past, present and future—an overview. Clim Change. 2005;70:9–29.
Williams IN, Torn MS, Riley WJ, Wehner MF. Impacts of climate extremes on gross primary production under global warming. Environ Res Lett. 2014;9:1–12.
Vasseur DA, DeLong JP, Gilbert B, Greig HS, Harley CDG, McCann KS, et al. Increased temperature variation poses a greater risk to species than climate warming. Proc Biol Sci. 2014;281:20132612–2.
Kremer CT, Fey SB, Arellano AA, Vasseur DA. Gradual plasticity alters population dynamics in variable environments: thermal acclimation in the green alga Chlamydomonas reinhartdii. Proc Biol Sci. 2018;285:20171942–9.
Rasconi S, Winter K, Kainz MJ. Temperature increase and fluctuation induce phytoplankton biodiversity loss—evidence from a multi-seasonal mesocosm experiment. Ecol Evol. 2017;7:2936–46.
Kremer CT, Klausmeier CA. Species packing in eco-evolutionary models of seasonally fluctuating environments. Ecol Lett. 2017;20:1158–68.
Schaum C-E, Rost B, Collins S. Environmental stability affects phenotypic evolution in a globally distributed marine picoplankton. ISME J. 2016;10:75–84.
Schaum C-E, Buckling A, Smirnoff N, Studholme DJ, Yvon-Durocher G. Environmental fluctuations accelerate molecular evolution of thermal tolerance in a marine diatom. Nat Commun. 2018;9:1719. https://doi.org/10.1038/s41467-018-03906-5.
Qu P, Fu F-X, Kling J, Huh M, Wang X, Hutchins DA. Distinct responses of Trichodesmium to a thermally-variable environment as a function of phosphorus availability. Front Microbiol. 2019;10:1282. https://doi.org/10.3389/fmicb.2019.01282.
Mantyla AW, Bograd SJ, Venrick EL. Patterns and controls of chlorophyll-a and primary productivity cycles in the Southern California Bight. J Mar Syst. 2008;73:48–60.
Nezlin NP, Sutula MA, Stumpf RP, Sengupta A. Phytoplankton blooms detected by SeaWiFS along the central and southern California coast. J Geophys Res. 2012;117:308–17.
Fuhrman JA, Hewson I, Schwalbach MS, Steele JA, Brown MV, Naeem S. Annually reoccurring bacterial communities are predictable from ocean conditions. Proc Natl Acad Sci USA. 2006;103:13104–9.
Countway PD, Vigil PD, Schnetzer A, Moorthi SD, Caron DA. Seasonal analysis of protistan community structure and diversity at the USC Microbial Observatory (San Pedro Channel, North Pacific Ocean). Limnol Oceanogr. 2010;55:2381–96.
Bruland KW, Rue EL, Smith GJ. Iron and macronutrients in California coastal upwelling regimes: Implications for diatom blooms. Limnol Oceanogr. 2001;46:1661–74.
Sunda W, Price N, Morel F, Andersen R. Trace metal metal ion buffers. Algal Culturing Techniques: Burlington, MA; 2005, p. 35–3.
Bernhardt JR, Sunday JM, Thompson PL, O’Connor MI. Nonlinear averaging of thermal experience predicts population growth rates in a thermally variable environment. Proc Biol Sci. 2018;285:20181076–10.
Fu F-X, Warner ME, Zhang Y, Feng Y, Hutchins DA. Effects of increased temperature and CO2 on photosynthesis, growth, and elemental ratios in marine Synechococcus and Prochlorococcus (Cyanobacteria). J Phycol. 2007;43:485–96.
Hutchins DA, DiTullio GR, Zhang Y, Bruland KW. An iron limitation mosaic in the California upwelling regime. Limnol Oceanogr. 1998;43:1037–54.
JD Strickland, TR Parsons. A practical handbook of seawater analysis. In: Stevenson JC, editor. Ottawa, Canada: Department of Fisheries and the Environment Fisheries and Marine Service Scientific Information and Publications Branch; 2012. p. 65–70.
Solórzano L, Sharp JH. Determination of total dissolved phosphorus and particulate phosphorus in natural waters. Limnol Oceanogr. 1980;25:754–8.
Xu K, Fu F-X, Hutchins DA. Comparative responses of two dominant Antarctic phytoplankton taxa to interactions between ocean acidification, warming, irradiance, and iron availability. Limnol Oceanogr. 2014;59:1919–31.
Parada AE, Needham DM, Fuhrman JA. Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ Microbiol. 2016;18:1403–14.
Needham DM, Fuhrman JA. Pronounced daily succession of phytoplankton, archaea and bacteria following a spring bloom. Nat Microbiol. 2016;1:1–7.
Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Meth. 2016;13:581–3.
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2012;41(D1):D590–6.
Decelle J, Romac S, Stern RF, Bendif EM, Zingone A, Audic S, et al. PhytoREF: a reference database of the plastidial 16S rRNA gene of photosynthetic eukaryotes with curated taxonomy. Mol Ecol Resour. 2015;15:1435–45.
Guillou L, Bachar D, Audic S, Bass D, Berney C, Bittner L, et al. The Protist Ribosomal Reference database (PR2): a catalog of unicellular eukaryote small sub-unit rRNA sequences with curated taxonomy. Nucleic Acids Res. 2012;41(D1):D597–D604.
Prokopowich CD, Gregory TR, Crease TJ. The correlation between rDNA copy number and genome size in eukaryotes. Genome. 2003;46:48–50.
Green BR. Chloroplast genomes of photosynthetic eukaryotes. Plant J. 2011;66:34–44.
Mizrahi-Man O, Davenport ER, Gilad Y. Taxonomic classification of bacterial 16S rRNA genes using short sequencing reads: evaluation of effective study designs. PLoS One. 2013;8:e53608–14.
R Core Team. R: A language and environment for statistical computing. R J 2018.
Racine J. RStudio: a platform-independent IDE for R and Sweave. J Appl Econ. 2012;27:167–72.
McMurdie PJ, Holmes S. Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE. 2013;8:e61217–11.
Oksanen J, Guillaume B, Friendly M, Kindt R, Legendre P, McGlinn D, et al. Vegan: a community ecology. R package version 2018; p. 1–297.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:31–21.
Kartal B, Van Niftrik L, Rattray J, Van De Vossenberg JL, Schmid MC, Sinninghe Damsté J, Jetten MS, Strous M. Candidatus ‘Brocadia fulgida’: an autofluorescent anaerobic ammonium oxidizing bacterium. FEMS Microbiol Ecol. 2008;63:46–55.
Dąbek P. et al. Towards a multigene phylogeny of the Cymatosiraceae (Bacillariophyta, Mediophyceae) I: novel taxa within the subfamily Cymatosiroideae based on molecular and morphological data. J Phycol. 2017;53:342–60.
Kuwata A, Yamada K, Ichinomiya M, Yoshikawa S, Tragin M, Vaulot D. et al. Bolidophyceae, a sister picoplanktonic group of diatoms—a review. Front Mar Sci. 2018;5:257–17.
Grossart H-P, Levold F, Allgaier M, Simon M, Brinkhoff T. Marine diatom species harbour distinct bacterial communities. Environ Microbiol. 2005;7:860–73.
Sohn JH, Lee J-H, Yi H, Chun J, Bae KS, Ahn T-Y, et al. Kordia algicida gen. nov., sp. nov., an algicidal bacterium isolated from red tide. Int J Syst Evol Microbiol. 2004;54(Pt 3):675–80.
Tont SA. Variability of diatom species populations: from days to years. J Mar Res. 1987;45:985–1006.
Schnetzer A, Miller PE, Schaffner RA, Stauffer BA, Jones BH, Weisberg SB, et al. Blooms of Pseudo-nitzschia and domoic acid in the San Pedro Channel and Los Angeles harbor areas of the Southern California Bight, 2003–2004. Harmful Algae. 2007;6:372–87.
Lemonnier H, Lantoine F, Courties C, Guillebault D, Nézan E, Chomérat N. et al. Dynamics of phytoplankton communities in eutrophying tropical shrimp ponds affected by vibriosis. Mar Pollut Bull. 2016;110:449–59.
Martinez EA. Sensitivity of marine ciliates (Protozoa, Ciliophora) to high thermal stress. Estuar Coast Mar Sci. 1980;10:369–IN1.
Sittenfeld A. Characterization of a photosynthetic Euglena strain isolated from an acidic hot mud pool of a volcanic area of Costa Rica. FEMS Microbiol Ecol. 2002;42:151–61.
Thomas MK, Aranguren-Gassis M, Kremer CT, Gould MR, Anderson K, Klausmeier CA, et al. Temperature-nutrient interactions exacerbate sensitivity to warming in phytoplankton. Glob. Chang Biol. 2017;23:3269–80.
Jiang H-B, Fu F-X, Rivero-Calle S, Levine NM, Sañudo-Wilhelmy SA, Qu P-P, et al. Ocean warming alleviates iron limitation of marine nitrogen fixation. Nat Clim Change. 2018;8:1–5.
Wernberg T, Smale DA, Tuya F, Thomsen MS, Langlois TJ, de Bettignies T, et al. An extreme climatic event alters marine ecosystem structure in a global biodiversity hotspot. Nat Clim Chang. 2012;5:1–5.
Siegle MR, Taylor EB, O’Connor MI. Prior heat accumulation reduces survival during subsequent experimental heat waves. J Exp Mar Bio Ecol. 2018;501:109–17.
Acknowledgements
Thanks to Troy Gunderson, Elaina Graham, Babak Hassanzadeh and the USC Wrigley Institute for Environmental Studies for help with logistics and analyses. Funding was provided by National Science Foundation awards OCE 1538525 and OCE 1638804 to FF and DAH.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kling, J.D., Lee, M.D., Fu, F. et al. Transient exposure to novel high temperatures reshapes coastal phytoplankton communities. ISME J 14, 413–424 (2020). https://doi.org/10.1038/s41396-019-0525-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41396-019-0525-6
This article is cited by
-
Symbiotic UCYN-A strains co-occurred with El Niño, relaxed upwelling, and varied eukaryotes over 10 years off Southern California
ISME Communications (2023)
-
Highly-resolved interannual phytoplankton community dynamics of the coastal Northwest Atlantic
ISME Communications (2022)
-
Environmental stability impacts the differential sensitivity of marine microbiomes to increases in temperature and acidity
The ISME Journal (2021)