Abstract
Background
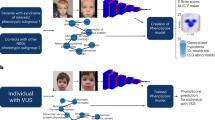
Congenital Central Hypoventilation Syndrome (CCHS) has devastating consequences if not diagnosed promptly. Despite identification of the disease-defining gene PHOX2B and a facial phenotype, CCHS remains underdiagnosed. This study aimed to incorporate automated techniques on facial photos to screen for CCHS in a diverse pediatric cohort to improve early case identification and assess a facial phenotype–PHOX2B genotype relationship.
Methods
Facial photos of children and young adults with CCHS were control-matched by age, sex, race/ethnicity. After validating landmarks, principal component analysis (PCA) was applied with logistic regression (LR) for feature attribution and machine learning models for subject classification and assessment by PHOX2B pathovariant.
Results
Gradient-based feature attribution confirmed a subtle facial phenotype and models were successful in classifying CCHS: neural network performed best (median sensitivity 90% (IQR 84%, 95%)) on 179 clinical photos (versus LR and XGBoost, both 85% (IQR 75–76%, 90%)). Outcomes were comparable stratified by PHOX2B genotype and with the addition of publicly available CCHS photos (n = 104) using PCA and LR (sensitivity 83–89% (IQR 67–76%, 92–100%).
Conclusions
Utilizing facial features, findings suggest an automated, accessible classifier may be used to screen for CCHS in children with the phenotype and support providers to seek PHOX2B testing to improve the diagnostics.
Impact
-
Facial landmarking and principal component analysis on a diverse pediatric and young adult cohort with PHOX2B pathovariants delineated a distinct, subtle CCHS facial phenotype.
-
Automated, low-cost machine learning models can detect a CCHS facial phenotype with a high sensitivity in screening to ultimately refer for disease-defining PHOX2B testing, potentially addressing gaps in disease underdiagnosis and allow for critical, timely intervention.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 14 print issues and online access
$259.00 per year
only $18.50 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Data availability
The clinical photos are HIPAA-protected and unavailable. The control photos in this project are publicly available through the UTKFace dataset and Dlib facial landmarking software is additionally open source. The publicly available photos with CCHS, code, and instruction may be available upon request.
References
Zhou, A. et al. Paired-like homeobox gene (PHOX2B) nonpolyalanine repeat expansion mutations (NPARMs): genotype–phenotype correlation in congenital central hypoventilation syndrome (CCHS). Genet. Med. 23, 1656–1663 (2021).
Weese-Mayer, D. E. et al. Congenital Central Hypoventilation Syndrome. Gene Reviews (eds Adam, M. P. et al) (University of Washington, 2021).
Jennings, L. J. et al. Variable human phenotype associated with novel deletions of the PHOX2B gene. Pediatr. Pulmonol. 47, 153–161 (2012).
Weese-Mayer, D. et al. An official ATS clinical policy statement: congenital central hypoventilation syndrome: genetic basis, diagnosis, and management. Am. J. Respir. Crit. Care Med. 181, 626 (2010).
Slattery, S. M. et al. Transitional care and clinical management of adolescents, young adults, and suspected new adult patients with congenital central hypoventilation syndrome. Clin. Auton. Res. 33, 231–249 (2022).
Trang, H. et al. Guidelines for diagnosis and management of congenital central hypoventilation syndrome. Orphanet J. Rare Dis. 15, 252–252 (2020).
Bachetti, T. & Ceccherini, I. Causative and common PHOX2B variants define a broad phenotypic spectrum. Clin. Genet. 97, 103–113 (2020).
Ogata, T. et al. Neurodevelopmental outcome and respiratory management of congenital central hypoventilation syndrome: a retrospective study. BMC Pediatr. 20, 342–342 (2020).
Todd, E. S. et al. Facial phenotype in children and young adults with PHOX2B –determined congenital central hypoventilation syndrome: quantitative pattern of dysmorphology. Pediatr. Res. 59, 39–45 (2006).
Solomon, B. D. et al. Perspectives on the future of dysmorphology. Am. J. Med. Genet. 191(Pt A), 659–671 (2023).
King, D. Dlib-ml: a machine learning toolkit. J. Mach. Learn. Res. 10, 1755–1758 (2009).
Ting, J., Song, C., Huang, H. & Tian, T. A comprehensive dataset for machine-learning-based lip-reading algorithm. Proc. Comput. Sci. 199, 1444–1449 (2022).
Li, X., Luo, J., Duan, C., Zhi, Y. & Yin, P. Real-time detection of fatigue driving based on face recognition. J. Phys. 1802, 22044 (2021).
Yang, J., Adu, J., Chen, H., Zhang, J. & Tang, J. A facial expression recongnition method based on Dlib, RI-LBP and ResNet. J. Phys. 1634, 12080 (2020).
Zhou, H., Chen, P. & Shen, W. A multi-view face recognition system based on cascade face detector and improved Dlib. In Proc. SPIE 10609, MIPPR 2017: Pattern Recognition and Computer Vision 1060908 (SPIE, 2018).
Boyko, N., Basystiuk, O. & Shakhovska, N. Performance evaluation and comparison of software for face recognition, based on Dlib and Opencv Library. In 2018 IEEE Second International Conference on Data Stream Mining & Processing (DSMP) 478–482 (IEEE, 2018).
Hennocq, Q., Khonsari, R. H., Benoît, V., Rio, M. & Garcelon, N. Computational diagnostic methods on 2D photographs: a review of the literature. J. Stomatol. Oral Maxillofac. Surg. 122, e71–e75 (2021).
Gurovich, Y. et al. Identifying facial phenotypes of genetic disorders using deep learning. Nat. Med. 25, 60–64 (2019).
Mahwish, N., Saherawala, B. A. & Jhancy, M. Clinical decision making in dysmorphology- emerging role of artificial intelligence. Br. J. Healthc. Med. Res. 9, 366–374 (2022).
Attallah, O. A deep learning-based diagnostic tool for identifying various diseases via facial images. Digital Health 8, 20552076221124432 (2022).
Mensah, M. A., Ott, C.-E., Horn, D. & Pantel, J. T. A machine learning-based screening tool for genetic syndromes in children. Lancet Digital Health 4, e295 (2022).
Wang, J. et al. Multiple genetic syndromes recognition based on a deep learning framework and cross-loss training. IEEE Access 10, 117084–117092 (2022).
Porras, A. R., Rosenbaum, K., Tor-Diez, C., Summar, M. & Linguraru, M. G. Development and evaluation of a machine learning-based point-of-care screening tool for genetic syndromes in children: a multinational retrospective study. Lancet 3, e635–e643 (2021).
Centers for Disease Control and Prevention. 2021 Child Development (Centers for Disease Control and Prevention, 2021).
Zhang, Z., Song, Y. & Qi, H. Age progression/regression by conditional adversarial autoencoder. Preprint at https://arxiv.org/abs/1702.08423. (2017).
Zhang, Z. & Song Y. UTKFace. Large Scale Face Dataset. https://susanqq.github.io/UTKFace/. (2017).
King, D. E. Dlib-models. https://github.com/davisking/dlib-models. (2002).
Chawla, N. V., Bowyer, K. W., Hall, L. O. & Kegelmeyer, W. P. SMOTE: synthetic minority over-sampling technique. J. Artif. Intell. Res. 16, 321–357 (2002).
Sirovich, L. & Kirby, M. Low-dimensional procedure for the characterization of human faces. J. Optical Soc. Am. A 4, 519 (1987).
Turk, M. & Pentland, A. Eigenfaces for recognition. J. Cogn. Neurosci. 3, 71–86 (1991).
Turk, M. A. & Pentland, A. P. Face recognition using eigenfaces. In Proc. 1991 IEEE Computer Society Conference on Computer Vision and Pattern Recognition 586–591 (IEEE, 1991).
Baehrens, D. et al. How to explain individual classification decisions. J. Mach. Learn. Res. 11, 1803–1831 (2010).
Chan, T. G. & Carlos, G. XGBoost: a scalable tree boosting system. In 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining 785–794 (ACM, 2016).
Paszke, A. et al. PyTorch: an imperative style, high-performance deep learning library. Adv. Neural Inf. Process. Syst. 32, 1–12 (2019).
Pedregosa, F. et al. Scikit-learn: machine learning in Python. J. Mach. Learn. Res. 12, 2825 (2011).
Slattery, S. M. et al. Ventilatory and orthostatic challenges reveal biomarkers for neurocognition in children and young adults with congenital central hypoventilation syndrome. Chest 163, 1555–1564 (2023).
Zelko, F. A. et al. Neurocognition as a biomarker in the rare autonomic disorders of CCHS and ROHHAD. Clin. Auton. Res. 33, 217–230 (2022).
Charnay, A. J. et al. Congenital central hypoventilation syndrome: neurocognition already reduced in preschool-aged children. Chest 149, 809 (2016).
Chen, S. et al. Development of a computer-aided tool for the pattern recognition of facial features in diagnosing Turner syndrome: comparison of diagnostic accuracy with clinical workers. Sci. Rep. 8, 9317 (2018).
Hennocq, Q. et al. An automatic facial landmarking for children with rare diseases. Am. J. Med. Genet. 191(Pt A), 1210–1221 (2023).
Kruszka, P. et al. 22q11.2 deletion syndrome in diverse populations: 22q11.2 Deletion Syndrome. Am. J. Med. Genet. 173(Pt A), 879–888 (2017).
Čaplovičová, M. et al. Modeling age‐specific facial development in Williams–Beuren‐, Noonan‐, and 22q11.2 deletion syndromes in cohorts of Czech patients aged 3–18 years: a cross‐sectional three‐dimensional geometric morphometry analysis of their facial gestalt. Am. J. Med. Genet. 176(Pt A), 2604–2613 (2018).
Liehr, T. et al. Next generation phenotyping in Emanuel and Pallister-Killian syndrome using computer-aided facial dysmorphology analysis of 2D photos. Clin. Genet. 93, 378–381 (2018).
McCradden, M. D., Joshi, S., Mazwi, M. & Anderson, J. A. Ethical limitations of algorithmic fairness solutions in health care machine learning. Lancet Digital Health 2, e221–e223 (2020).
McCradden, M. D. & Chad, L. Screening for facial differences worldwide: equity and ethics. Lancet Digital Health 3, e615–e616 (2021).
Pantel, J. T. et al. Efficiency of computer-aided facial phenotyping (DeepGestalt) in individuals with and without a genetic syndrome: diagnostic accuracy study. J. Med. Internet Res. 22, e19263 (2020).
Hong, D. et al. Genetic syndromes screening by facial recognition technology: VGG-16 screening model construction and evaluation. Orphanet J. Rare Dis. 16, 344 (2021).
Su, Z. et al. Deep learning-based facial image analysis in medical research: a systematic review protocol. BMJ Open 11, e047549 (2021).
Dingemans, A. J. M., de Vries, B. B. A., Vissers, L. E. L. M., van Gerven, M. A. J. & Hinne, M. Comparing facial feature extraction methods in the diagnosis of rare genetic syndromes. Preprint at medRxiv https://doi.org/10.1101/2022.08.26.22279217. (2022).
Zhao, Q. et al. Digital facial dysmorphology for genetic screening: Hierarchical constrained local model using ICA. Med. Image Anal. 18, 699–710 (2014).
Dingemans, A. J. M. et al. PhenoScore quantifies phenotypic variation for rare genetic diseases by combining facial analysis with other clinical features using a machine-learning framework. Nat. Genet. 55, 1598–1607 (2023).
Acknowledgements
We are grateful for the introductory discussions held with Pavel Prusakov, PhD; Bimal Chaudhari, MD, MPH; George El-Ferzli, MD; Sven Bambach, PhD; and Steve Rust, PhD.
Funding
This study is funded in part by the Chicago Community Trust Foundation PHOX2B Patent Fund.
Author information
Authors and Affiliations
Contributions
All authors have met authorship requirements. Specifically, S.M.S., J.W., A.M., C.Z., N.E., S.S., J.J.B., T.M.S., D.D. and D.E.W.-M. had substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data. S.M.S., J.W., A.M., C.Z., N.E., S.S., J.J.B., C.M.R., I.K., T.M.S., D.D. and D.E.W.-M. drafted the article or revised it critically for intellectual content. S.M.S., J.W., A.M., C.Z., N.E., S.S., J.J.B., C.M.R., I.K., T.M.S., D.D. and D.E.W.-M. provided final approval of this version to be published.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Consent statement
Obtaining clinical photos of patients with CCHS is included in the standardized inpatient evaluation at the Center for Autonomic Medicine in Pediatrics at Ann & Robert H. Lurie Children’s Hospital of Chicago. For the four included in Fig. 2 of the manuscript, a separate and explicit consent statement was obtained from legal representatives of each child.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Slattery, S.M., Wilkinson, J., Mittal, A. et al. Computer-aided diagnostic screen for Congenital Central Hypoventilation Syndrome with facial phenotype. Pediatr Res (2024). https://doi.org/10.1038/s41390-023-02990-8
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41390-023-02990-8