Abstract
Background
Dilated cardiomyopathy (DCM) in children is often associated with poor morbidity and mortality and exhibits distinct pathological entities from those of adult DCM. Owing to the limited number of patients and the lack of a good animal model, the molecular mechanisms underlying pediatric DCM remain poorly understood. The purpose of this study is to establish an animal model of neonatal DCM and identify early progression factors.
Methods
Cardiac phenotypes and comprehensive gene expression profiles in homozygous ΔK210 knock-in (TNNT2ΔK210/ΔK210) mice were analyzed and compared to TNNT2+/ΔK210 and wild-type mice at 0 days and 1 week of age.
Results
Immediately after birth, the cardiac weight in TNNT2ΔK210/ΔK210 mice was already increased compared to that in TNNT2+/ΔK210 and wild-type mice. Echocardiographic examination of 0-day-old and 1-week-old TNNT2ΔK210/ΔK210 mice revealed similar phenotypes of pediatric DCM. In addition, several genes were significantly upregulated in the ventricular tissues of TNNT2ΔK210/ΔK210 mice, and the KEGG PATHWAY analysis revealed several important pathways such as cancer and focal adhesion that might be associated with the pathogenesis and development of DCM.
Conclusions
TNNT2ΔK210/ΔK210 mice have already developed DCM at birth, indicating that they should be an excellent animal model to identify early progression factors of DCM.
Impact
-
TNNT2ΔK210/ΔK210 mice are excellent animal model for DCM.
-
TNNT2ΔK210/ΔK210 mice are excellent animal model to identify early progression factors of DCM.
-
KEGG PATHWAY analysis revealed that several important pathways such as cancer and focal adhesion might be associated with the pathogenesis and development of neonatal DCM.
Similar content being viewed by others
Introduction
Dilated cardiomyopathy (DCM) is characterized by cardiac dilation and pump failure. It is the most common type of cardiomyopathy and heart failure in children and is diagnosed in 0.57 of every 100,000 children per year in the United States.1,2 Among DCM patients who are diagnosed at or before 10 years of age, 20% are diagnosed during the neonatal period (<4 weeks old) and another 65% are diagnosed in the first year of life.3 Approximately half of all children with DCM either die or undergo heart transplantation within the first 5 years.2 In 20–45% of pediatric patients with DCM, on the other hand, cardiac function can be normalized.4 Recent studies have indicated that pediatric and adult DCM are distinct pathological entities.5,6 For example, the gene profiles and responses to stress in pediatric DCM are different from those in adult DCM.4 Despite tremendous efforts to improve available treatment protocols, outcomes of children with DCM have improved little and heart transplantation is almost the only therapeutic strategy. The reasons for our lack of progress in this regard include the limited number of patients and the lack of a good animal model for pediatric DCM. A useful animal model is therefore highly desirable as it will enable us to more thoroughly investigate the molecular mechanisms underlying pediatric DCM.
Of the DCM patients who are diagnosed at or before 10 years of age, 15% are considered to have familial DCM.3 Pediatric DCM is often associated with mutations in sarcomeric and cytoskeletal genes. Cardiac troponin T (TNNT2) exon 13 lysine deletions (ΔK210) often result in a rapid progression of heart failure or sudden cardiac death at a young age.7,8,9 To clarify the causal mechanisms of DCM, TNNT2 ΔK210 knock-in mice have been generated; these exhibit phenotypes similar to those in human DCM, including premature death.10 Homozygous or heterozygous ΔK210 knock-in (TNNT2ΔK210/ΔK210 or TNNT2+/ΔK210) mice start to die at approximately 10 or 30 days after birth, respectively.10 Only 20% of TNNT2ΔK210/ΔK210 mice can survive longer than 4 months. Therefore, ΔK210 knock-in mice are considered to be an excellent animal model of pediatric DCM. In spite of their usefulness, however, the phenotypes of ΔK210 knock-in mice have not been extensively analyzed during the neonatal and weaning periods. The purpose of the present study is to investigate these early phenotypes of TNNT2ΔK210/ΔK210 mice from birth and to explore the molecular mechanisms underlying the poor prognosis of TNNT2ΔK210/ΔK210 mice.
Materials and methods
Animals
Wild-type, TNNT2+/ΔK210, and TNNT2ΔK210/ΔK210 mice were used throughout this study and were examined either immediately after birth (0 days old) or 1 week after birth (1 week old). To obtain tissue samples, all animals were anesthetized with isoflurane, and cervical dislocation was performed after loss of pain reflex. Thereafter, the heart was immediately excised and weighed. A portion of each animal’s ventricular tissues was frozen in liquid nitrogen as a sample for gene expression analysis and another portion was soaked in 10% formaldehyde neutral buffer solution (Nacalai Tesque, Inc., Kyoto, Japan) for histological examination. The animal experiments were performed in accordance with the NIH guidelines (Guide for the Care and Use of Laboratory Animals). All animal studies were approved by the Institutional Animal Care and Use Committee of The Jikei University.
Echocardiography
To evaluate the global morphology and function of the heart, echocardiography was performed at 0 days and 1 week of age in three groups of mice. All mice were sedated with 1.5% isoflurane using an up to 70-MHz transducer from the Vevo 3100 Imaging System (FUJIFILM VisualSonic Inc., Toronto, Canada). Measurements were performed using the analysis software Vevo LAB (FUJIFILM VisualSonic Inc.). Interventricular septum (IVS), left ventricular (LV) diameter at end diastole, LV diameter at end systole, posterior LV wall thickness (PWT), ejection fraction (EF), and LV end-diastolic volume were measured at the papillary muscle level of the LV using a short-axis view.
Histological examination
The excised ventricular tissues of wild-type, TNNT2+/ΔK210, and TNNT2ΔK210/ΔK210 mice at 0 days and 1 week of age were subjected to hematoxylin–eosin (HE) and Masson trichrome staining. Each sample was observed under a light microscope at 10 times magnification. The photographed images were converted to combined images using BZ-9000 (KEYENCE, Osaka, Japan) to estimate the fibrosis area in Masson trichrome staining as previously described.11
DNA microarray analysis
Total RNAs were extracted from ventricular tissue in wild-type and TNNT2ΔK210/ΔK210 mice at 0 days and 1 week of age using TRIZOL reagent (Thermo Fisher Scientific, Waltham, MA, USA). Three ventricular samples from each group were combined for comparison. RNA purity and integrity were evaluated using an ND-1000 Spectrophotometer (NanoDrop, Wilmington, DE, USA) and an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA). Total RNA labeling and hybridization were performed using the Agilent One-Color Microarray-Based Gene Expression Analysis protocol (Agilent Technologies, V 6.5, 2010). Briefly, 100 ng of total RNA was labeled with Cy3-dCTP. The labeled cRNAs were purified using an RNAeasy Mini Kit (Qiagen, Hilden, Germany). The concentration and specific activity of the labeled cRNAs (pmol Cy3/μg cRNA) were measured using the NanoDrop ND-1000 Spectrophotometer. Hybridization was performed according to the supplier’s instructions (Agilent Technologies, V 6.5, 2010). Six hundred nanograms of each labeled cRNA was fragmented by adding 5 μl 10× blocking agent and 1 μl of 25× fragmentation buffer, and then heated at 60 °C for 30 min. Finally, 25 μl 2× GE hybridization buffer was added to dilute the labeled cRNA. Forty microliters of hybridization solution was dispensed into the gasket slide and assembled to the Agilent SurePrint G3 Mouse GE 8 × 60K Microarrays (Agilent). The slides were incubated for 17 h at 65 °C in an Agilent hybridization oven and then washed at room temperature according to the Agilent One-Color Microarray-Based Gene Expression Analysis protocol (Agilent Technologies, V 6.5, 2010). Hybridization signals were immediately scanned with an Agilent Microarray Scanner D (Agilent Technologies). Microarray results were extracted using the Agilent Feature Extraction software v11.0 (Agilent Technologies). Array probes that had Flag A in their samples were filtered out. Selected gProcessedSignal value was transformed by logarithm and normalized according to the quantile method. Statistical significance of the expression data was determined using local pooled error test and fold change in which the null hypothesis was that no difference exists among groups. False discovery rate was controlled by adjusting the p value by means of the Benjamini–Hochberg algorithm. For a differentially expressed gene set, hierarchical cluster analysis was performed using complete linkage and Euclidean distance as a measure of similarity. Gene Enrichment and Functional Annotation analysis for significant probe list was performed using the Kyoto Encyclopedia of Genes and Genomes (KEGG) PATHWAY database (www.genome.jp/kegg/).12 All data analysis and visualization of DEGs was conducted using R 3.1.2 (www.r-project.org). The complete data set of the DNA microarray is available in the Gene Expression Omnibus database (accession number: GSE133052).
Real-time PCR analysis
Extraction of total RNA from ventricular tissues from wild-type, TNNT2+/ΔK210, and TNNT2ΔK210/ΔK210 mice at 0 days and 1 week of age, generation of cDNA, and reverse transcription PCR (RT-PCR) analysis were performed as described previously.11 Natriuretic peptide type A (ANF), brain natriuretic factor (BNF), connective tissue growth factors (CTGFs), glial cell line-derived neurotrophic factor (Gdnf), secreted phosphoprotein 1 (Spp1), protein phosphatase 1, regulatory (inhibitor) subunit 12B (Ppp1r12b), sarcolipin (SLN), phospholamban (PLN), calsequestrin 1 (CSQ1) and 2 (CSQ2), sarcoplasmic reticulum (SR)/endoplasmic reticulum Ca2+-ATPase 2a (SERCA2a), ryanodine receptor type 2 (RyR2) troponin T type A (TnT A) and E (TnT E), cardiac troponin I (cTnI), slow skeletal type troponin I (ssTnI), cardiac troponin C (cTnC), and cardiac tropomyosin (cTM) expression levels in the ventricular tissues were measured in the wild-type, TNNT2+/ΔK210, and TNNT2ΔK210/ΔK210 mice at 0 days and 1 week of age using real-time PCR. The primer nucleotide sequences are shown in Supplementary Table S1. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) expression level was quantitated as an internal control.
Western blot analysis
Protein was extracted from ventricular tissues from wild-type and TNNT2ΔK210/ΔK210 mice at 0 days and 1 week of age. Ventricular tissues were lysed in RIPA buffer containing phenylmethylsulfonyl fluoride, protease inhibitor, and sodium orthovanadate (Santa Cruz Biotechnology, CA, USA). Samples were centrifuges at 15,000 rpm for 10 min, and the protein concentration of supernatants was determined using the BCA Protein Assay Kit (TAKARA Bio Inc., Shiga, Japan). An aliquot of the extracted protein solution was mixed with sample buffer solution (Nacalai Tesque, Inc., Kyoto, Japan), and the mixture was heated at 95 °C for 5 min. Thirty micrograms of protein were separated on 4–20% gradient sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS PAGE) gels (Bio-Rad, CA, USA) and electrically transferred from the gels to a polyvinylidene difluoride membrane (Merck Millipore, MA, USA). After blocking with 5% skim milk in TBS with 0.05% Tween-20, primary antibodies were applied to the membrane overnight at 4 °C. After washing, the membranes were incubated with horseradish peroxidase-conjugated secondary antibody. The signals were detected using the West Dura Extended Duration Substrate (Thermo Fisher Scientific, MA, USA). The band intensities of the target proteins were analyzed using the NIH Image J software and normalized by the band intensity of GAPDH (Cell Signaling Technology, MA, USA). The primary antibodies used in this study were as follows: anti-col4a5 (Genetex, CA, USA), anti-Birc5 (Cell Signaling Technology, MA, USA), anti-Lpar3 (Abcam, Tokyo, Japan), anti-Fos (Cell Signaling Technology, MA, USA), anti-troponin I (Cell Signaling Technology, MA, USA), anti-phospho-troponin I (Ser23/34) (Cell Signaling Technology, MA, USA), and anti-GAPDH (Cell Signaling Technology, MA, USA). For sarcomeric protein detection, 10 μg of protein were separated on 10% SDS PAGE gels (Bio-Rad, CA, USA) and the SYPRO Ruby Protein Gel Stain (Thermo Fisher Scientific, MA, USA) was used according to the manufacturer’s protocol.
Statistical analysis
Results are presented as the mean ± standard error of the mean (SEM). Surgical and echocardiographic findings and various mRNA expression levels from real-time PCR were compared between these groups using analysis of variance followed by post hoc Tukey’s multiple comparison test. p < 0.05 was considered significant.
Results
Neonatal TNNT2 ΔK210/ΔK210 mice displayed a dilated cardiomyopathic phenotype
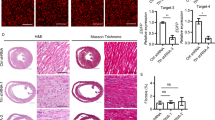
Neonatal TNNT2ΔK210/ΔK210 mice displayed a significant increase in heart/body weight ratio when compared with wild-type mice and TNNT2+/ΔK210 mice at 0 days of age (Table 1), whereas the heart/body weight ratio was not different between wild-type mice and TNNT2+/ΔK210 mice at either 0 days or 1 week of age. At neither age was there any significant difference in body weight among the three groups. After Masson trichrome staining, representative horizontal sections of the heart at the papillary muscle level are shown in Fig. 1a. There was no difference in the incidence or distribution of fibrotic areas and disarray of cardiomyocytes among the three groups at birth or 1 week after birth (Fig. 1b), although previous studies have reported that marked interstitial fibrosis developed in the ventricles of 6-week-old TNNT2ΔK210/ΔK210 mice.10 After HE staining, myofibril organization also remained normal in TNNT2+/ΔK210 and TNNT2ΔK210/ΔK210 mice at birth and 1 week after birth (Supplementary Fig. S1). Echocardiographic examination of 0-day-old neonatal mice revealed that no significant difference in LV EF (Fig. 2f), an index of systolic function, among the three groups, whereas LV end-diastolic dimension (Fig. 2c) and volume (Fig. 2e) were significantly increased in TNNT2ΔK210/ΔK210 mice as compared with wild-type mice. Echocardiographic examination of 1-week-old neonatal mice also revealed that LV end-diastolic dimension (Fig. 2c) and volume (Fig. 2e) were significantly increased in TNNT2ΔK210/ΔK210 mice as compared with wild-type and TNNT2+/ΔK210 mice, and that LV EF (Fig. 2f) was significantly reduced in TNNT2ΔK210/ΔK210 mice as compared with wild-type and TNNT2+/ΔK210 mice. IVS thickness (Fig. 2a) was significantly thinner in 1-week-old TNNT2ΔK210/ΔK210 mice than in wild-type and TNNT2+/ΔK210 mice, whereas no significant differences were detected in LV PWT among the three groups at either 0 days or 1 week of age (Fig. 2b).
a Representative histological sections were stained with Masson trichrome staining in the heart at the papillary muscle level of WT, TNNT2+/ΔK210, and TNNT2ΔK210/ΔK210 mice at 0 days and 1 week of age. Scale bars are 500 μm. b Quantification of fibrotic areas in the hearts of WT (n = 7), TNNT2+/ΔK210 (n = 8) and TNNT2ΔK210/ΔK210 (n = 5) mice at 0 days and 1 week of age. Data are shown as mean ± SEM.
Echocardiographic examination of 0-day-old (n = 3–4 per group) and 1-week-old (n = 5–6 per group) neonatal WT, TNNT2+/ΔK210, and TNNT2ΔK210/ΔK210 mice. a Interventricular septal (IVS) thickness, b posterior left ventricular wall thickness (PWT), c left ventricular diameter at end diastole (LVDd), d left ventricular diameter at end systole (LVDs), e left ventricular end-diastolic volume (LVEDV), and f ejection fraction. Data are shown as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001 by ANOVA followed by post hoc Tukey’s multiple comparison test.
DNA microarray analyses revealed distinct transcriptional profiles of TNNT2 ΔK210/ΔK210 mice
We performed microarray analysis with pooled LV tissues from wild-type and TNNT2ΔK210/ΔK210 mice at 0 days (n = 2 for each genotype) and 1 week of age (n = 2 for each genotype). The degree of similarity in the transcriptional profiles between the wild-type and TNNT2ΔK210/ΔK210 mice was calculated, and the profiles of gene expression were grouped into two distinct clusters: a 0-day-old cluster and a 1-week-old cluster (Supplementary Fig. S2), suggesting that the effect of development on the gene profiles is much greater than the effect of genotype. Nevertheless, we found that 12 gene probes at 0 days of age (Table 2) and 53 gene probes at 1 week of age (Table 3) were significantly upregulated (fold change = ≥2.0 and p value < 0.05) in the pooled ventricular tissues of TNNT2ΔK210/ΔK210 mice as compared with those of wild-type mice. Two genes (calsequestrin 1 and contactin 2) were significantly upregulated in TNNT2ΔK210/ΔK210 mice of both ages. On the other hand, we found that 20 gene probes at 0 days of age (Table 4) and 30 gene probes at 1 week of age (Table 5) were significantly downregulated (fold change = ≤−2.0 and p value < 0.05) in the pooled ventricular tissues of TNNT2ΔK210/ΔK210 mice as compared with those of wild-type mice. Three genes (secreted phosphoprotein 1, Rab7b, and AK041267) were significantly downregulated in TNNT2ΔK210/ΔK210 mice of both ages.
To analyze the interaction networks and reaction networks of the genes that have distinct expression levels in TNNT2ΔK210/ΔK210 mice as compared to wild-type mice, we performed the KEGG PATHWAY analysis using the KEGG database.12 There was significant (p < 0.05) overrepresentation of genes involved in five KEGG pathways in TNNT2ΔK210/ΔK210 mice as compared with wild-type mice and TNNT2ΔK210/ΔK210 mice at 1 week of age, namely, cancer, focal adhesion, cardiac muscle contraction, adrenergic signaling in cardiomyocytes, and phosphatidylinositol 3-kinase-Akt signaling pathways (Fig. 3).
RT-PCR analysis verified the expression patterns of DNA microarray analyses of TNNT2 ΔK210/ΔK210 mice
To verify the results of the microarray, we performed quantitative RT-PCR for ANF, BNF, CTGF, Gdnf, Spp1, and Ppp1r12b. We found that ANF levels was highly expressed in the ventricles of 1-week-old TNNT2ΔK210/ΔK210 mice compared to wild-type and TNNT2+/ΔK210 mice; at birth, in contrast, the expression levels of ANF mRNA were not different among the three groups (Fig. 4a). The expression levels of BNF (Fig. 4b) and CTGF (Fig. 4c), on the other hand, were not different among the three groups at either 0 days or 1 week of age. Furthermore, we found that the expression levels of Gdnf were significantly higher in TNNT2ΔK210/ΔK210 mice than in wild-type mice at both 0 days and 1 week (Fig. 4d). The expression levels of Spp1 (Fig. 4e) and Ppp1r12b (Fig. 4f) were significantly lower in TNNT2ΔK210/ΔK210 mice than in wild-type mice at 0 days; at 1 week of age, Spp1 levels remained significantly lower in TNNT2ΔK210/ΔK210 mice, while Ppp1r12b levels were comparable between TNNT2ΔK210/ΔK210 and wild-type mice. These data supported our DNA microarray results.
Quantification of RT-PCR products for a ANF (n > 6 per group), b BNF (n > 6 per group), c CTGF (n > 4 per group), d Gdnf (n > 3 per group), e Spp1 (n > 3 per group), and f Ppp1r12b (n > 3 per group) in ventricular tissues from wild-type (WT), TNNT2+/ΔK210, and TNNT2ΔK210/ΔK210 mice at 0 days and 1 week of age. Data are shown as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001 by ANOVA followed by post hoc Tukey’s multiple comparison test.
Western blot analysis verified the expression patterns of DNA microarray analysis of TNNT2 ΔK210/ΔK210 mice
To determine whether TNNT2 K210 deletion induces alterations in the expression levels of other sarcomeric proteins, we investigated total protein using SYPRO Ruby Protein Gel Stain. Sarcomeric protein expression levels were not significantly different between wild-type and TNNT2ΔK210/ΔK210 mice at either 0 days or 1 week of age (Supplementary Fig. S3). To verify the results of the DNA microarray analysis, we performed western blot analysis for Col4a5, Birc5, Lpar3, and Fos. The protein expression level of Birc5 was higher in TNNT2ΔK210/ΔK210 mice than in wild-type mice, while Col4a5, Lpar3, and Fos expression levels were comparable between TNNT2ΔK210/ΔK210 and wild-type mice at 0 days of age (Fig. 5a). The protein expression levels of Col4a5, Lpar3, and Birc5 were found to be upregulated, and Fos expression levels were found to be downregulated in TNNT2ΔK210/ΔK210 mice as compared to wild-type mice at 1 week of age (Fig. 5b, c). These results are consistent with the results of DNA microarray analysis shown in Tables 3 and 5.
Protein expression levels of Col4a5, Birc5, Lpar3, cTnI, and pcTnI in a pooled left ventricular tissues from wild-type and TNNT2ΔK210/ΔK210 mice at 0 days and b left ventricular tissues from wild-type and TNNT2ΔK210/ΔK210 mice at 1 week of age. c Quantification of protein expression levels of Col4a5, Birc5, and Lpar3 and relative phosphorylation levels of cTnI in wild-type and TNNT2ΔK210/ΔK210 mice at 1 week of age. Data are shown as mean ± SEM. *p < 0.05 by ANOVA followed by post hoc Tukey’s multiple comparison test.
The changes in transcription profiles of SR-related genes in TNNT2 ΔK210/ΔK210 mice
Next, we examined the expression levels of genes related to the SR because SR dysfunction is known to be one of the earliest insults in heart failure. We found that SLN (Supplementary Fig. S4A) and CSQ1 (Supplementary Fig. S4C) were significantly upregulated in TNNT2ΔK210/ΔK210 mice at 1 week of age, which is consistent with our DNA microarray data. In addition, the expression levels of CSQ2 were significantly increased in TNNT2ΔK210/ΔK210 mice at 1 week of age compared to those in wild-type and TNNT2+/ΔK210 mice (Supplementary Fig. S4D). With regard to PLN (Supplementary Fig. S4B), SERCA2a (Supplementary Fig. S4E), and RyR2 (Supplementary Fig. S4F) mRNAs, in contrast, there was no difference among the three groups at either 0 days or 1 week of age.
The changes in transcription profiles of troponin-related genes in TNNT2 ΔK210/ΔK210 mice
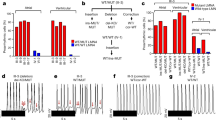
We also investigated whether troponin T amino acid mutation (ΔK210) may change the composition of troponin T isoforms and troponin T-related myofibril proteins in TNNT2ΔK210/ΔK210 mice during the early neonatal period. Our examination of the expression levels of isoforms of troponin T (A-type: fatal form or E-type: adult form), isoforms of troponin I (cardiac type or slow skeletal type), cardiac/slow skeletal troponin C, and tropomyosin 1 revealed no significant differences among the three groups (Fig. 6). To verify the effects of troponin mutation on sarcomere protein phosphorylation, we examined the phosphorylation levels of cardiac troponin I. The phosphorylation levels of cardiac troponin I were not significantly different between TNNT2ΔK210/ΔK210 mice and wild-type mice at 1 week of age (Fig. 4b, c).
Discussion
The present study showed that the mean ventricular mass and LV end-diastolic dimension and volume of TNNT2ΔK210/ΔK210 mice has already increased at birth and that these mice rapidly develop cardiac dysfunction within 1 week after birth, suggesting that TNNT2ΔK210/ΔK210 mice could be an excellent animal model to investigate the molecular mechanisms of neonatal DCM, especially as this genetic mutation is also found in human DCM. After demonstrating this model’s usefulness, we used it to explore the mechanisms of early progression in neonatal DCM.
DNA microarray analyses revealed that the gene expression profiles of TNNT2ΔK210/ΔK210 mice at birth (0 days old) were slightly different from those of wild-type mice and that these differences were even larger 1 week after birth. The expression levels of ANF mRNA, a biomarker of cardiac stress and dysfunction, were not different among the three groups at birth but were significantly increased in TNNT2ΔK210/ΔK210 mice at 1 week of age. Consistent with the result, echocardiographic data also indicated in impaired cardiac function in TNNT2ΔK210/ΔK210 mice at the first week of life, but not at birth, as shown in Fig. 2f. Therefore, we assume that the week following birth is a critical period for the initiation of DCM progression of TNNT2ΔK210/ΔK210 mice, and we are keenly interested in the genes whose expression levels changed during the first week of life (see Tables 3 and 5). KEGG PATHWAY analysis revealed that these genes are involved in five important pathways that might be associated with the pathogenesis and development of DCM. Although the finding of cancer and focal adhesion pathways is of particular interest, we do not have a definite answer how these pathways play a role in neonatal DCM. However, several previous studies have also demonstrated that cancer13,14 or focal adhesion pathways15 would be involved in DCM. Six of the genes are involved in cancer-related pathways: four genes (Col4a5, Lpar3, Birc5 (survivin40), and Cks1b) were significantly increased and two genes (Fos and Ptch2) were significantly decreased in TNNT2ΔK210/ΔK210 mice at age 1 week. Huang et al. have suggested that Fos is likewise significantly downregulated in human DCM samples and that it plays an important role in the progression of heart failure as a hub gene for signaling pathways,16 which is consistent with the present results. On the other hand, Windak et al. have demonstrated that Fos is dispensable for adaptive cardiac hypertrophy using pressure overload-induced cardiac hypertrophy in mice and targeted deletion of Fos in cardiomyocytes.17 These data indicate that downregulation of Fos is involved in a pathological pathway leading to the development of heart failure but not in a physiological pathway leading to hypertrophy.
Birc5, also known as survivin, inhibits apoptosis and is highly expressed in common human cancers. Birc5 is a critical factor in determining the total number and proliferation of cardiomyocytes and is indispensable for postnatal cardiac development.18 Although the expression levels of Birc5 decrease with postnatal cardiac development,19 Birc5 is upregulated in the peri-infarct zone and remote myocardium in human myocardial infarction20 and elevated in human18 and rat heart failure,21 which is consistent with the present findings. Thus the role of Birc5 in the early progression of neonatal DCM is worth further investigation.
The role of the other four cancer-related-pathway genes in the development of cardiomyopathy has not yet been investigated in detail. The COL4A5 gene, which codes non-fibrillar collagen, was reportedly increased in 23 human LV tissue samples from DCM patients.22 That report, however, is the only one to date that suggests that the upregulation of COL4A5 mRNA may be associated with the fibrotic process in DCM. In the present study, the expression levels of COL4A5 mRNA were increased up to 3.7-fold in TNNT2ΔK210/ΔK210 mice at 1 week of age; it was therefore among the top three most upregulated genes in this study. Although we did not find fibrosis in TNNT2ΔK210/ΔK210 mice at 1 week of age, we have previously shown the progression of fibrosis in TNNT2ΔK210/ΔK210 mice at 8 weeks of age.10 The upregulation of COL4A5 mRNA may proceed the fibrotic changes. Further investigation is needed to qualify this assumption. Another gene whose expression level changed during the first week of life in TNNT2ΔK210/ΔK210 mice was Lpar3, an embryonic isoform of lysophospholipid G protein-coupled receptor via which small signaling lipids regulate diverse physiological and pathological processes.23 The upregulation of Lpar3 (2.1 fold) in TNNT2ΔK210/ΔK210 mice may reflect the activation of a fetal gene program.
Although SR dysfunction is known to be one of the earliest insults in heart failure, the expression levels of key molecules affecting the cardiac SR, including PLN, SERCA2a, and RyR2, remained unchanged in the first week of life. In our previous unpublished examinations, the expression of SERCA2a mRNA was decreased after 4 weeks of age in TNNT2ΔK210/ΔK210 mice. SLN and CSQ1, in contrast, were significantly upregulated within 1 week after birth in TNNT2ΔK210/ΔK210 mice. Interestingly, SLN and CSQ1 are not usually expressed in the ventricles. Rather, SLN is highly expressed in the atrium and skeletal muscles, although the induction of SLN in the ventricles is known to happen under specific conditions.24,25 CSQ1 is a skeletal type of calsequestrin isoform and the induction of CSQ1 in the ventricles has not been reported. Although we do not know the mechanism by which these skeletal muscle types of SR genes are induced in the neonatal period in TNNT2ΔK210/ΔK210 mice, an epigenetic program might be involved in this phenomenon. Since SLN is a negative regulator of SERCA2a and SERCA1, and since CSQ1 inhibits RyR1 at a physiological luminal calcium concentration,26 we assume that the upregulation of SLN and CSQ1 may inhibit calcium cycling in the hearts of TNNT2ΔK210/ΔK210 mice, which would play a role in the early progression of cardiac dysfunction in TNNT2ΔK210/ΔK210 mice at 1 week of age.
Because cardiac troponin T isoforms are known to switch from the fetal type (TnT A) to the adult type (TnT E) during the neonatal period,27,28 and because their Ca2+ sensitivities differ with regard to myofibrillar force development,29,30 we had hypothesized that the timing of the isoform switch might affect the onset of cardiac dysfunction in TNNT2ΔK210/ΔK210 mice. However, we did not find any differences among the three groups in the expression levels of troponin T isoforms, other troponin-related genes, or phosphorylation levels of cardiac troponin I. This finding indicates that the composition of troponin T and associated thin-filament genes is not affected in the progression of cardiac dysfunction.
In conclusion, the mean ventricular mass was already increased at birth in TNNT2ΔK210/ΔK210 mice, and their cardiac function became impaired at age 1 week. Our KEGG PATHWAY analysis revealed that several important pathways such as cancer and focal adhesion might be associated with the pathogenesis and development of neonatal DCM. The present study indicated that TNNT2ΔK210/ΔK210 mice should be an excellent animal model to identify early progression factors of DCM.
References
Lipshultz, S. E. et al. The incidence of pediatric cardiomyopathy in two regions of the United States. N. Engl. J. Med. 348, 1647–1655 (2003).
Towbin, J. A. et al. Incidence, causes, and outcomes of dilated cardiomyopathy in children. JAMA 296, 1867–1876 (2006).
Daubeney, P. E. et al. Clinical features and outcomes of childhood dilated cardiomyopathy: results from a national population-based study. Circulation 114, 2671–2678 (2006).
Lee, T. M. et al. Pediatric cardiomyopathies. Circ. Res. 121, 855–873 (2017).
Patel, M. D. et al. Pediatric and adult dilated cardiomyopathy represent distinct pathological entities. JCI Insight 2, e94382 (2017).
Tatman, P. D. et al. Pediatric dilated cardiomyopathy hearts display a unique gene expression profile. JCI Insight 2, e94249 (2017).
Hanson, E. L. et al. Cardiac troponin T lysine 210 deletion in a family with dilated cardiomyopathy. J. Card. Fail. 8, 28–32 (2002).
Martins, E. et al. Familial dilated cardiomyopathy with troponin T K210del mutation. Rev. Port. Cardiol. 25, 295–300 (2006).
Mogensen, J. et al. Severe disease expression of cardiac troponin C and T mutations in patients with idiopathic dilated cardiomyopathy. J. Am. Coll. Cardiol. 44, 2033–2040 (2004).
Du, C. K. et al. Knock-in mouse model of dilated cardiomyopathy caused by troponin mutation. Circ. Res. 101, 185–194 (2007).
Kusakari, Y. et al. Impairment of excitation-contraction coupling in right ventricular hypertrophied muscle with fibrosis induced by pulmonary artery banding. PLoS ONE 12, e0169564 (2017).
Kanehisa, M. et al. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res 34, D354–D357 (2006).
Chen, S. N. et al. DNA damage response/TP53 pathway is activated and contributes to the pathogenesis of dilated cardiomyopathy associated with LMNA (Lamin A/C) mutations. Circ. Res. 124, 856–873 (2019).
Hou, N. et al. Activation of Yap1/Taz signaling in ischemic heart disease and dilated cardiomyopathy. Exp. Mol. Pathol. 103, 267–275 (2017).
Xiao, J., Li, F., Yang, Q., Zeng, X. F. & Ke, Z. P. Co-expresssion analysis provides important module and pathways of human dilated cardiomyopathy. J. Cell. Physiol. 235, 494–503 (2020).
Huang, H. et al. Identification of potential gene interactions in heart failure caused by idiopathic dilated cardiomyopathy. Med. Sci. Monit. 24, 7697–7709 (2018).
Windak, R. et al. The AP-1 transcription factor c-Jun prevents stress-imposed maladaptive remodeling of the heart. PLoS ONE 8, e73294 (2013).
Levkau, B. et al. Survivin determines cardiac function by controlling total cardiomyocyte number. Circulation 117, 1583–1593 (2008).
Sheng, L. et al. Downregulation of Survivin contributes to cell-cycle arrest during postnatal cardiac development in a severe spinal muscular atrophy mouse model. Hum. Mol. Genet. 27, 486–498 (2018).
Santini, D. et al. Surviving acute myocardial infarction: survivin expression in viable cardiomyocytes after infarction. J. Clin. Pathol. 57, 1321–1324 (2004).
Abbate, A. et al. Myocardial expression of survivin, an apoptosis inhibitor, in aging and heart failure. An experimental study in the spontaneously hypertensive rat. Int. J. Cardiol. 111, 371–376 (2006).
Gil-Cayuela, C. et al. New altered non-fibrillar collagens in human dilated cardiomyopathy: role in the remodeling process. PLoS ONE 11, e0168130 (2016).
Wang, F. et al. Developmental changes in lysophospholipid receptor expression in rodent heart from near-term fetus to adult. Mol. Biol. Rep. 39, 9075–9084 (2012).
Pashmforoush, M. et al. Nkx2-5 pathways and congenital heart disease; loss of ventricular myocyte lineage specification leads to progressive cardiomyopathy and complete heart block. Cell 117, 373–386 (2004).
Voit, A. et al. Reducing sarcolipin expression mitigates Duchenne muscular dystrophy and associated cardiomyopathy in mice. Nat. Commun. 8, 1068 (2017).
Beard, N. A., Sakowska, M. M., Dulhunty, A. F. & Laver, D. R. Calsequestrin is an inhibitor of skeletal muscle ryanodine receptor calcium release channels. Biophys. J. 82, 310–320 (2002).
Wang, Q., Reiter, R. S., Huang, Q. Q., Jin, J. P. & Lin, J. J. Comparative studies on the expression patterns of three troponin T genes during mouse development. Anat. Rec. 263, 72–84 (2001).
Wei, B. & Jin, J. P. TNNT1, TNNT2, and TNNT3: isoform genes, regulation, and structure-function relationships. Gene 582, 1–13 (2016).
Pinto, J. R. et al. Fetal cardiac troponin isoforms rescue the increased Ca2+ sensitivity produced by a novel double deletion in cardiac troponin T linked to restrictive cardiomyopathy: a clinical, genetic, and functional approach. J. Biol. Chem. 286, 20901–20912 (2011).
Venkatraman, G., Gomes, A. V., Kerrick, W. G. & Potter, J. D. Characterization of troponin T dilated cardiomyopathy mutations in the fetal troponin isoform. J. Biol. Chem. 280, 17584–17592 (2005).
Acknowledgements
The authors acknowledge Ms. Naoko Tomizawa for her technical support of this research. This work was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to J.T., S. Morimoto, S. Minamisawa), the Vehicle Racing Commemorative Foundation (S. Minamisawa), the Uehara Memorial Foundation (S. Minamisawa), the Descente and Ishimoto Memorial Foundation for Promotion of Sports Science (J.T.), The Jikei University Graduate Student Research Grant (Y.F., S.B.), and the Miyata Cardiology Research Promotion Foundation (S. Minamisawa).
Author information
Authors and Affiliations
Contributions
J.T., T.F., S.B., and Y.F. conceived and performed the experiments. J.T., S. Morimoto, and S. Minamisawa conducted and supervised the experiments. S. Morimoto and S. Minamisawa designed the experiments. J.T. and S. Minamisawa wrote the manuscript, and all authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Tanihata, J., Fujii, T., Baba, S. et al. Troponin T amino acid mutation (ΔK210) knock-in mice as a neonatal dilated cardiomyopathy model. Pediatr Res 89, 846–857 (2021). https://doi.org/10.1038/s41390-020-1016-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41390-020-1016-1