Abstract
Background
Neuroblastoma is the commonest extracranial solid cancer for neonates. Long non-coding RNA cancer susceptibility 11 (CASC11) is corroborated as carcinogen in several tumors. But its role in neonatal neuroblastoma is poorly defined.
Methods
Expression levels of CASC11, miR-676-3p, and NOL4L mRNA were analyzed by qRT-PCR in cells and tissues. Kaplan–Meier analysis was used to measure and analyze the survival time of patients with high/low CASC11. Neonatal neuroblastoma cell proliferation was reflected through colony-formation assay and CCK-8. Transwell assay was designed for detection of migratory and invasive capacities of neonatal neuroblastoma cells. Wound-healing assay was used for monitoring neuroblastoma cell migration. RNA pull-down, luciferase reporter, and RIP assays were utilized to identify the relationship between CASC11, miR-676-3p, and NOL4L on the basis of bioinformatics tools.
Results
Highly expressed CASC11 was observed in neonatal neuroblastoma tissues and cells. High level of CASC11 indicated unsatisfactory survival of neonatal neuroblastoma patients. CASC11 depletion inhibited cell proliferation and invasiveness. CASC11 was a molecular sponge to release NOL4L from miR-676-3p inhibition in tumor cells. Upregulation of NOL4L abated the suppressed cell proliferation and invasiveness due to CASC11 downregulation.
Conclusion
CASC11 sequestered miR-676-3p from NOL4L to facilitate neonatal neuroblastoma progression, hinting a CASC11-mediated therapeutic target for neonatal neuroblastoma.
Similar content being viewed by others
Introduction
Neuroblastoma has been fairly common in children and led to 15% of all childhood deaths from tumor.1,2 Neuroblastoma is identified exclusively as a pediatric malignancy since 90% patients are aged <10 years.3 In addition, neonatal neuroblastoma is attributed to >20% of neonatal malignancies.4 Despite the continuous improvement of management strategies, the survival rate of children diagnosed as neonatal neuroblastoma is frustrating over the decades. An in-depth study of the neonatal neuroblastoma pathogenesis is imperatively needed for the discovery of new treating methods.5
Long non-coding RNAs (lncRNAs) are referred to a set of non-translated transcripts typically >200 nucleotides in length and possess essential impacts of modulating gene expression.6 Functionally, a repertoire of biological processes including proliferation and metastasis are tightly associated with lncRNAs.7 For example, lncRNA DANCR potentiates nasopharyngeal carcinoma metastasis through the interaction with NF90/NF45 complex.8 LncRNA SLCO4A1-AS1 contributes to the activation of β-catenin-dependent Wnt pathway to facilitate growth and metastasis in colorectal cancer.9 In neuroblastoma progression, a wealth of lncRNAs has been evidenced as crucial modulators in recent years.10,11 LncRNA FOXD3-AS1 is documented to inhibit neuroblastoma progression through repressing the activation of CTCF.12 LncRNA cancer susceptibility 11 (CASC11) is located on 8q24.21 and previously reported to be a carcinogenic factor in colorectal cancer,13 hepatocellular carcinoma,14 and so on. Till now, whether CASC11 is functionally implicated in neuroblastoma initiation and development has not been certified.
In this work, we illuminated the increased level of CASC11 in neuroblastoma tissues and cells and unfavorable survival caused by its overexpression was disclosed. Moreover, reduced CASC11 hampered neuroblastoma cell proliferation and invasiveness. Mechanically, CASC11 potentiated neuroblastoma cell proliferation and invasiveness through liberating nucleolar protein 4 like (NOL4L) from microRNA-676-3p.
Materials and methods
Clinical specimens
Neonatal neuroblastoma patients enrolled in The First Hospital of Jilin University supplied the related tissues in this work including 42 neonatal neuroblastoma tissues and 42 normal tissue samples. Ethics Committee of the aforementioned hospital has offered the approval for the conduction of our study. Prior to the performance of our study, all the enrolled patients agreed to sign on the letter of consent.
Cell culture
SK-N-AS and NB-1 were the neonatal neuroblastoma cells, and the cell line of human retinal-pigmented epithelial immortalized by telomerase reverse transcriptase (hTERT-RPE1) was the control cell line, which were all gained from American Type Culture Collection (ATCC, Manassas, VA, USA). Dulbecco’s Modified Eagle’s Medium (Invitrogen) was for SK-N-AS cell culture supplemented with 10% fetal bovine serum (FBS; Invitrogen) and 0.1 mM Non-Essential Amino Acids (Invitrogen). NB-1 cells were maintained in mixed medium with 1:1 ratio of F12 medium (Invitrogen) and Minimum Essential Medium (Invitrogen, Carlsbad, CA, USA) containing 10% FBS (Invitrogen). hTERT-RPE1 cells were cultured with 5% circulating CO2 at 37 °C in RPMI-1640 medium (Life Technologies, USA) containing 10% FBS (Life Technologies, USA), penicillin (100 U/ml), and streptomycin (100 mg/ml) (Life Technologies).
Reagents and cell transfection
The forced expression of NOL4L was achieved by constructing PCR-amplified NOL4L sequence into pcDNA3.1 vector (Invitrogen) to establish pcDNA3.1/NOL4L overexpression plasmids. Short hairpin RNAs (shRNAs) against CASC11 (sh/CASC11#1/2/3) and controls sh/NC, miR-676-3p mimics and miR-676-3p inhibitor and their controls NC mimics, and NC inhibitor were attained from GenePharma Co. Ltd (Suzhou, China). Abiding by the protocols of suppliers, we transfected these plasmids into SK-N-AS and NB-1 cells using Lipofectamine 2000 reagent (Invitrogen).
RNA extraction and real-time PCR
Total RNA isolation from SK-N-AS and NB-1 or hTERT-RPE1 cells or tissues was achieved using Trizol reagent (Invitrogen, Carlsbad, CA, USA). Applying random primers and Oligo dT primer, lncRNA CASC11 and NOL4L mRNA were reverse transcribed, respectively. Quantitative reverse transcriptase PCR (qRT-PCR) was conducted following the SYBR Green PCR Kit (Toyobo, Osaka, Japan) protocol on Applied Biosystems 7300 real-time PCR system (Applied Biosystems, Foster City, CA, USA). U6 acted as the internal control of miR-676-3p. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as an internal control for CASC11 and NOL4L mRNA. The sequences of primers are displayed as below: CASC11 forward, AGAGCGCTGGAGAAAATGCT, reverse TTTGTGTCTCGGTTCTCCATAG; NOL4L forward, TCAAGTCCTGCCGTCGCATGAA, reverse TTGGCTGCCTTTCTTGTCTCGC; GAPDH forward, AGCCACATCGCTCAGACAC, reverse GCCCAATACGACCAAATCC.
Cell Counting Kit-8 (CCK-8) assay
Referring to the instructions of a Cell Counting Kit-8 Kit (Dojindo Molecular Technologies, Rockville, MD, USA), SK-N-AS and NB-1 cell proliferative capacity was examined 24 h after transfection. Generally, cell suspensions (100 μl) after being dispensed into 96-well plates were pre-incubated for 24 h. CCK-8 solution (10 μl) was added at the indicated time points into each well and underwent incubation for another 3 h. Epoch Microplate Spectrophotometer (Bio Tek, Winooski, VT) detected the cell absorbance (450 nm) to reflect cell viability.
Colony-formation assay
SK-N-AS and NB-1 cells following transfection were placed into culture dishes (60 mm) and maintained for about 14 days in the required complete medium. Next, absolute ethyl alcohol was utilized for SK-N-AS and NB-1 fixation for 15 min, and SK-N-AS and NB-1 staining was completed for 10 min by crystal violet solution (0.1%, Sigma-Aldrich, St. Louis, MO, USA). Eventually, positive colonies (>50 cells) were counted.
Transwell migration and invasion assay
To probe SK-N-AS and NB-1 cell migration, 5 × 104 SK-N-AS and NB-1 after 48 h transfection in serum-free media were dispersed into the upper chamber (8-μm pore size, Millipore). For the invasion assays, 1 × 105 cells prepared in serum-free media were transferred into the upper chamber coated with Matrigel (Sigma-Aldrich). Media supplemented with 10% FBS were added into the lower chamber. Twenty-four hours later, SK-N-AS and NB-1 cells that migrated or invaded through the membrane were first stained by 0.1% crystal violet, and optical microscope (Olympus, Tokyo, Japan) counted the stained cells.
Wound-healing assay
A total of 3 × 105 SK-N-AS and NB-1 cells seeded in 6-well plates were cultured overnight and transduced with several plasmids. When the confluence reached 85%, the cell layer was scratched by sterile plastic tip, followed by culture medium washing. Then SK-N-AS and NB-1 cells were maintained for 48 h in serum-free medium containing 1% FBS. At different time points, a microscope was used for acquiring images of the plates. Digimizer software system was applied for wound distance measurement.
Biotin pull-down assay
Total protein extraction from SK-N-AS and NB-1 cells were accomplished with lysis buffer and incubated overnight with CASC11 probe labeled with biotin or controls at 4 °C. Afterwards, streptavidin-coated magnetic beads (Invitrogen) were used for incubation with cell lysates for 2 h at 25 °C. Then microRNAs (miRNAs) that bound to CASC11 were analyzed by qRT-PCR.
RNA immunoprecipitation (RIP) assay
RIP experiments were executed as per the manufacturer’s protocol of the EZ-Magna RIP Kit (Millipore, Billerica, MA) using the Ago2 antibody (Abcam, Cambridge, MA, USA). Lysed SK-N-AS and NB-1 cells by complete RIP lysis buffer was subjected to RIP buffer incubation at 4 °C containing magnetic beads coupled with Ago2 or control IgG antibody (Millipore) for 6 h. After the beads were washed and proteins in the complexes were removed with 0.1% sodium dodecyl sulfate/0.5 mg/ml Proteinase K, NanoDrop spectrophotometer (Thermo Scientific) was utilized for RNA concentration measurement and bioanalyzer (Agilent, Santa Clara, CA) was for quality assessment. Immunoprecipitated RNAs was probed by qRT-PCR.
Luciferase reporter assay
SK-N-AS and NB-1 cells at 3 × 104 cells/well were dispersed in 24-well plates and settled overnight. The next day, reporter plasmids CASC11-WT/MUT and miR-676-3p mimics or NC mimics were co-transfected into SK-N-AS and NB-1 cells. Likewise, SK-N-AS and NB-1 cells were co-transfected with NOL4L-WT/MUT reporter plasmids and miR-676-3p mimics or NC mimics. Twenty-four hours later, the luciferase activity normalized against Renilla was monitored under the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA).
Cell cytoplasm/nucleus fraction isolation
Cytoplasmic and nuclear extracts from SK-N-AS and NB-1 cells were collected using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Scientific, Waltham, MA, USA). qRT-qPCR analysis was employed for the analysis of RNAs isolated from both the fractions. The levels of nuclear control (U1), cytoplasmic control (GAPDH), and CASC11 were assessed.
RNA fluorescence in situ hybridization (FISH) assay
U1 and CASC11 probes were synthesized and provided by Bersinbio Company (Guangzhou, China). SK-N-AS and NB-1 cell slides were first fixed for 20 min in 4% paraformaldehyde and subjected to protein K digestion for 10 min. After PBS washes of slides, hybridization reaction solution (20 μl) was added, followed by overnight hybridization. Then the slides were washed twice by 25% formamide/2× Saline Sodium Citrate. Finally, the slides were subjected to 4,6-diamidino-2-phenylindole staining and fluorescent signal measurement by Zeiss LSM800 confocal laser microscopy (Zeiss, Germany).
Statistical analysis
Statistical analyses were carried out using the Graphpad Prism 6 software. Data from three times repeated experiments were compared by either Student’s t test or analysis of variance (one way) when appropriate and mean ± standard deviation was calculated. Pearson’s correlation coefficient was designed for analyzing the association between CASC11 and miR-676-3p or NOL4L expression. Kaplan–Meier’s method was used for addressing the impacts of CASC11 on the survival of neonatal neuroblastoma patients. P < 0.05 was considered to be statistically significant.
Results
CASC11 upregulation predicts poor survival of neonatal neuroblastoma patients and is mainly distributed in the cytoplasm
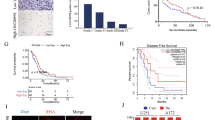
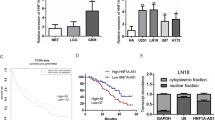
From the qRT-PCR analysis of neonatal neuroblastoma and normal tissues, we ascertained that CASC11 was a highly expressed lncRNA in neonatal neuroblastoma (n = 42), compared with 42 cases of normal samples (Fig. 1a). Forty-two patients were divided into two groups on the basis of CASC11 median value. By the analysis of Kaplan–Meier’s method, it testified that CASC11 upregulation was correlated with dreadful survival in neonatal neuroblastoma patients (Fig. 1b). In addition, elevated CASC11 was observed in SK-N-AS and NB-1 cells than in control hTERT-RPE1 cell line (Fig. 1c). Prior to the investigation of CASC11 function on neonatal neuroblastoma, we studied the subcellular distribution of CASC11 in different fractions. CASC11 was unveiled to preferentially exist in SK-N-AS and NB-1 cytoplasm (Fig. 1d). FISH analysis validated that CASC11 was dominantly located in the cytoplasm (Fig. 1e). Results showed that, as a cytoplasmic lncRNA, CASC11 elevation correlated with poor survival in neonatal neuroblastoma.
CASC11 upregulation predicts poor survival of neonatal neuroblastoma patients and is mainly distributed in cytoplasm. a Neonatal neuroblastoma tissues and normal samples were subjected to qRT-PCR analysis of CASC11 expression. b Survival time of patients with increased CASC11 was shortened than that of patients with low CASC11 expression. c Analysis of CASC11 level in neuroblastoma cells and control cell line. d Cell cytoplasm/nucleus fraction isolation with qRT-PCR indicated the presence of CASC11. e FISH assayed that the CASC11 was mainly in the cytoplasm. **P < 0.01
Knockdown of CASC11 hinders growth and invasiveness of neuroblastoma cells
To verify whether CASC11 was implicated in the regulation of neuroblastoma cells, SK-N-AS and NB-1 cells were transfected with shRNAs against CASC11 to interfere CASC11 expression (Fig. 2a). CCK-8 and colony formation illuminated that CASC11 knockdown by specific shRNAs led to cell viability inhibition of SK-N-AS and NB-1 cells (Fig. 2b, c). In addition, the migration and invasion abilities of SK-N-AS and NB-1 cells in the sh/CASC11-transfected group were declined in transwell assay (Fig. 2d). Wound-healing assay demonstrated the repressed SK-N-AS and NB-1 cell migration following CASC11 depletion (Fig. 2e). All data provided evidence for the growth-promoting and aggressiveness-facilitating role of CASC11 in neuroblastoma cells.
Knockdown of CASC11 hinders growth and invasiveness for neuroblastoma cells. a Depleted CASC11 level was found in neuroblastoma cells after transfection. b Neuroblastoma cells following CASC11 depletion was analyzed by CCK-8 assay. c Neuroblastoma cells with or without CASC11 depletion were subjected to colony-formation assay. d Transwell assay examined the invasiveness property of neuroblastoma cells following CASC11 suppression. e Wound-healing assay was for determining neuroblastoma cell migration after downregulating CASC11. **P < 0.01
CASC11 functions as miR-676-3p sponge to antagonize miR-676-3p-mediated NOL4L mRNA degradation
We sought to detail the mechanism by which CASC11 exerted in neuroblastoma. Cytoplasmic lncRNA is identified to sequester away miRNAs as competing endogenous ceRNAs and thereby augment mRNA expression.15,16 Using starbase (http://starbase.sysu.edu.cn/) and DIANA (http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php?r = site%2Ftools) softwares, we revealed three miRNAs (miR-6805-3p, miR-676-3p, and miR-2355-3p) that were interacting with CASC11 (Fig. 3a). Based on this prediction, we performed RNA pull-down assay. The results showed that miR-676-3p, not the other two (miR-6805-3p, miR-2355-3p), were enriched by CASC11 probe compared with control probe (Fig. 3b). Inhibited miR-676-3p level was found in neonatal neroblastoma tissues in comparison to normal specimen. Correlation analysis pointed out the inverse correlation between miR-676-3p and CASC11 expression in tumor tissues (Fig. 3c). Moreover, miRmap, RNA22, and microT predicted that miR-676-3p targeted NOL4L (Fig. 3d). NOL4L was highly expressed in cancer tissues relative to healthy controls and positively correlated with CASC11 expression in tumor tissues (Fig. 3e). Meanwhile, NOL4L was inversely correlated with miR-676-3p (Fig. 3e). The binding sites of NOL4L and CASC11 for the miR-676-3p binding are shown (Fig. 3f). RIP assay described that CASC11 and NOL4L interacted with miR-676-3p in an Ago2-dependent manner (Fig. 3g). Wild-type CASC11 or NOL4L and corresponding mutant reporters (miR-676-3p-binding site was mutated) were co-transfected with miR-676-3p mimics or NC mimics into SK-N-AS and NB-1 cells. We found that cells transfected with CASC11-WT and miR-676-3p mimics exhibited decreased luciferase activity. Likewise, miR-676-3p mimics led to the declined luciferase activity of NOL4L-WT but not the mutant one (Fig. 3h). CASC11 inhibition decreased NOL4L mRNA and protein while miR-676-3p inhibitor negated this impact (Fig. 3i). These data elucidated the existence of CASC11-NOL4L crosstalk through miR-676-3p-binding competition.
CASC11 functions as miR-676-3p sponge to antagonize miR-676-3p-mediated NOL4L mRNA degradation. a Three miRNAs that interacted with CASC11 were predicted by the DIANA and starbase software. b RNA pull-down detected the interaction between CASC11 and three predicted miRNAs. c miR-676-3p level was monitored in neonatal neuroblastoma tissues and control tissues; the correlation between miR-676-3p and CASC11 level was probed in tumor tissues. d NOL4L was unveiled by miRmap, RNA22, and microT to bind to miR-676-3p. e NOL4L expression in neonatal neuroblastoma tissues and control samples was examined, and it positively correlated with CASC11; correlation between miR-676-3p and NOL4L was probed in tumor tissues. f Predicted binding sites between CASC11, miR-676-3p, and NOL4L are exhibited. g CASC11, miR-676-3p, and NOL4L that bound to IgG or Ago2 were monitored by qRT-PCR after RIP. h Indicated cells after transfection with wild-type or mutant CASC11 or NOL4L reporters and miR-676-3p mimics or NC mimics were subjected to luciferase reporter assay. i NOL4L level of mRNA and protein in response to the downregulation of CASC11 and miR-676-3p was investigated. **P < 0.01. NS denotes no significant difference
CASC11 regulates neuroblastoma cell growth and migration through NOL4L
Subsequently, we tested whether CASC11 exerted its function through NOL4L. Then NOL4L was overexpressed, as ensured by qRT-PCR and western blot (Fig. 4a). Functionally, the proliferation reduction by CASC11 depletion was restored via the overexpression of NOL4L (Fig. 4b, c). Transwell assay found that NOL4L elevation abolished the anti-aggressiveness effect of CASC11 downregulation (Fig. 4d). Similarly, wound-healing assay unveiled that CASC11 inhibition-mediated suppression of cell migration was negated by NOL4L augmentation (Fig. 4e). These observations uncovered that CASC11 exerted its tumor-promoting function through upregulating NOL4L.
CASC11 regulates neuroblastoma cell growth and migration through NOL4L. a Overexpression efficacy of NOL4L was tested by qRT-PCR. b, c CCK-8 and colony formation were conducted in cells transfected with sh/NC, sh/CASC11#2, sh/CASC11#2+pcDNA3.1, and sh/CASC11#2+pcDNA3.1/NOL4L. d Cell migration and invasion was monitored. e Wound-healing assay was used for reflecting cell migration after the indicated treatments. *P < 0.05, **P < 0.01. NS denotes no significant difference
Discussion
Neuroblastoma is recognized as a life-threatening malignancy in young children and infants. During the pathogenesis of neuroblastoma, it was closely correlated with significantly altered lncRNAs and multiple processes were regulated by them.10,11 For example, highly expressed lncRNA CAI2 contributes to p16 overexpression and advanced-stage neuroblastoma.17 LncRNA MEG3, HCN3, and linc01105 affect neuroblastoma cell proliferation and apoptosis through the hypoxia-inducible factor-1α and p53 pathways.18 Besides, lncRNAs could also be conducive to prognostic biomarker identification. For example, lncRNA SNHG16 is positively associated with undesirable prognosis of pediatric neuroblastoma.19 However, lncRNAs that are functionally characterized or used as characteristic biomarkers were reported. CASC11 is proved as an oncogenic lncRNA in multiple diseases. For example, overexpression of CASC11 increases cancer cell resistance to chemotherapy in ovarian squamous cell carcinoma.20 It also aggravates the progression of colorectal cancer,13 gastric cancer,21 osteosarcoma,22 bladder cancer,23 and hepatocellular carcinoma.14 Its role in neonatal neuroblastoma has not been substantiated. In the present study, we first found CASC11 was overexpressed in both neonatal neuroblastoma tissues and cells, and its high expression was positively correlated with frustrating clinical outcomes. Moreover, it was first revealed that silencing CASC11 curbed neuroblastoma cell proliferation and invasiveness.
The regulation of lncRNAs functioning in tumors is partly dependent on its subcellular distribution. Generally speaking, cytoplasmic-enriched lncRNAs reduces the availability of miRNAs to target mRNAs through competitively binding to miRNA.24 CASC11 is reported to sponge miR-340-5p21 and miR-188-5p.25 In this study, CASC11 was preferentially presented in the cytoplasm of neuroblastoma cells. Besides it sponged miR-676-3p whose expression was negatively correlated with CASC11. Furthermore, CASC11 antagonized the inhibitory function of miR-676-3p on NOL4L, which was previously recorded to involve in acute myeloid leukemia.26 Previous work did not functionally identify NOL4L in tumors. In this study, we disclosed that highly expressed NOL4L antagonized the proliferation-suppressing or antimigration effect of CASC11 depletion in neonatal neuroblastoma. Mechanically, the interplay and the expression correlation between CASC11, miR-676-3p, and NOL4L were first observed in neonatal neuroblastoma.
In conclusion, it suggested that CASC11 was neonatal neuroblastoma-related oncogene, which promoted neuroblastoma progression through upregulating NOL4L by serving as a miR-676-3p sponge. Our observations will provide basic evidence for treating CASC11 as an effective target for neonatal neuroblastoma therapy.
References
Whittle, S. B. et al. Overview and recent advances in the treatment of neuroblastoma. Expert Rev. Anticancer Ther. 17, 369–386 (2017).
Ward, E., DeSantis, C., Robbins, A., Kohler, B. & Jemal, A. Childhood and adolescent cancer statistics, 2014. CA Cancer J. Clin. 64, 83–103 (2014).
Esiashvili, N., Goodman, M., Ward, K., Marcus, R. B. Jr & Johnstone, P. A. Neuroblastoma in adults: Incidence and survival analysis based on SEER data. Pediatr. Blood Cancer 49, 41–46 (2007).
Fisher, J. P. H. & Tweddle, D. A. Neonatal neuroblastoma. Semin. Fetal Neonatal Med. 17, 207–215 (2012).
Interiano, R. B. & Davidoff, A. M. Current management of neonatal neuroblastoma. Curr. Pediatr. Rev. 11, 179–187 (2015).
Bergmann, J. H. & Spector, D. L. Long non-coding RNAs: modulators of nuclear structure and function. Curr. Opin. Cell Biol. 26, 10–18 (2014).
Huarte, M. The emerging role of lncRNAs in cancer. Nat. Med 21, 1253–1261 (2015).
Wen, X. et al. Long non-coding RNA DANCR stabilizes HIF-1alpha and promotes metastasis by interacting with NF90/NF45 complex in nasopharyngeal carcinoma. Theranostics 8, 5676–5689 (2018).
Yu, J. et al. LncRNA SLCO4A1-AS1 facilitates growth and metastasis of colorectal cancer through beta-catenin-dependent Wnt pathway. J. Exp. Clin. Cancer Res. 37, 222 (2018).
Pandey, G. K. et al. The risk-associated long noncoding RNA NBAT-1 controls neuroblastoma progression by regulating cell proliferation and neuronal differentiation. Cancer Cell 26, 722–737 (2014).
Pichler, M. & Calin, G. A. Long noncoding RNA in neuroblastoma: new light on the (old) N-Myc story. J. Natl. Cancer Inst. 106, dju150 (2014).
Zhao, X. et al. Risk-associated long noncoding RNA FOXD3-AS1 inhibits neuroblastoma progression by repressing PARP1-mediated activation of CTCF. Mol. Ther. 26, 755–773 (2018).
Zhang, Z. et al. Long non-coding RNA CASC11 interacts with hnRNP-K and activates the WNT/beta-catenin pathway to promote growth and metastasis in colorectal cancer. Cancer Lett. 376, 62–73 (2016).
Han, Y., Chen, M., Wang, A. & Fan, X. STAT3-induced upregulation of lncRNA CASC11 promotes the cell migration, invasion and epithelial-mesenchymal transition in hepatocellular carcinoma by epigenetically silencing PTEN and activating PI3K/AKT signaling pathway. Biochem. Biophys. Res. Commun. 508, 472–479 (2019).
Salmena, L., Poliseno, L., Tay, Y., Kats, L. & Pandolfi, P. P. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 146, 353–358 (2011).
Sen, R., Ghosal, S., Das, S., Balti, S. & Chakrabarti, J. Competing endogenous RNA: the key to posttranscriptional regulation. ScientificWorldJournal 2014, 896206 (2014).
Barnhill, L. M. et al. High expression of CAI2, a 9p21-embedded long noncoding RNA, contributes to advanced-stage neuroblastoma. Cancer Res. 74, 3753–3763 (2014).
Tang, W., Dong, K., Li, K., Dong, R. & Zheng, S. MEG3, HCN3 and linc01105 influence the proliferation and apoptosis of neuroblastoma cells via the HIF-1alpha and p53 pathways. Sci. Rep. 6, 36268 (2016).
Yu, Y. et al. lncRNA SNHG16 is associated with proliferation and poor prognosis of pediatric neuroblastoma. Int. J. Oncol. 55, 93–102 (2019).
Shen, F. et al. Overexpression of CASC11 in ovarian squamous cell carcinoma mediates the development of cancer cell resistance to chemotherapy. Gene 710, 363–366 (2019).
Zhang, L. et al. LncRNA CASC11 promoted gastric cancer cell proliferation, migration and invasion in vitro by regulating cell cycle pathway. Cell Cycle 17, 1886–1900 (2018).
Song, K., Yuan, X., Li, G., Ma, M. & Sun, J. Long noncoding RNA CASC11 promotes osteosarcoma metastasis by suppressing degradation of snail mRNA. Am. J. Cancer Res. 9, 300–311 (2019).
Luo, H., Xu, C., Le, W., Ge, B. & Wang, T. lncRNA CASC11 promotes cancer cell proliferation in bladder cancer through miRNA-150. J. Cell. Biochem. 120, 13487–13493 (2019).
Schmitt, A. M. & Chang, H. Y. Long noncoding RNAs in cancer pathways. Cancer Cell 29, 452–463 (2016).
Cheng, N. et al. LncRNA CASC11 promotes cancer cell proliferation in hepatocellular carcinoma by inhibiting miRNA-188-5p. Biosci. Rep. 39, BSR20190251 (2019).
Guastadisegni, M. C. et al. CBFA2T2 and C20orf112: two novel fusion partners of RUNX1 in acute myeloid leukemia. Leukemia 24, 1516–1519 (2010).
Author information
Authors and Affiliations
Contributions
Z.Y. and J.Z.: manuscript editing, review, and data analysis. J.H.: figures’ preparation.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Yu, Z., Zhang, J. & Han, J. Silencing CASC11 curbs neonatal neuroblastoma progression through modulating microRNA-676-3p/nucleolar protein 4 like (NOL4L) axis. Pediatr Res 87, 662–668 (2020). https://doi.org/10.1038/s41390-019-0625-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41390-019-0625-z
This article is cited by
-
Role of non-coding RNAs in neuroblastoma
Cancer Gene Therapy (2023)
-
Circ_0132817 facilitates cell proliferation, migration, invasion and glycolysis by regulating the miR-432-5p/NOL4L axis in neuroblastoma
Experimental Brain Research (2021)