Abstract
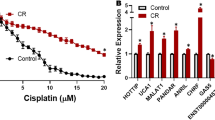
Cisplatin resistance is a major cause of therapeutic failure in patients with high-grade serous ovarian cancer (HGSOC). Long noncoding RNAs (lncRNAs) have emerged as key regulators of human cancers; however, their modes of action in HGSOC remain largely unknown. Here, we provide evidence to demonstrate that lncRNA Platinum sensitivity-related LncRNA from Ascites-Derived Exosomes (PLADE) transmitted by ascites exosomes enhance platinum sensitivity in HGSOC. PLADE exhibited significantly decreased expression in ascites exosomes and tumor tissues, as well as in the corresponding metastatic tumors from patients with HGSOC cisplatin-resistance. Moreover, HGSOC patients with higher PLADE expression levels exhibited longer progression-free survival. Gain- and loss-of-function studies have revealed that PLADE promotes cisplatin sensitivity by suppressing cell proliferation, migration and invasion, and enhancing apoptosis in vitro and in vivo. Furthermore, the functions of PLADE in increasing cisplatin sensitivity were proven to be transferred by exosomes to the cultured recipient cells and to the adjacent tumor tissues in mouse models. Mechanistically, PLADE binds to and downregulates heterogeneous nuclear ribonucleoprotein D (HNRNPD) by VHL-mediated ubiquitination, thus inducing an increased amount of RNA: DNA hybrids (R-loop) and DNA damage, consequently promoting cisplatin sensitivity in HGSOC. Collectively, these results shed light on the understanding of the vital roles of long noncoding RNAs in cancers.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
All original data generated from Next Generation Sequencing (NGS) in this study have been deposited in the Gene Expression Omnibus (GEO) database under the accession number GSE247337. All study data and materials are included in the article and/or supporting information.
References
Lheureux S, Gourley C, Vergote I, Oza AM. Epithelial ovarian cancer. Lancet. 2019;393:1240–53.
Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72:7–33.
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer J Clin. 2021;71:209–49.
La Vecchia C. Ovarian cancer: epidemiology and risk factors. Eur J Cancer Prev. 2017;26:55–62.
O’Malley DM. New therapies for ovarian cancer. J Natl Compr Canc Netw. 2019;17:619–21.
Kuroki L, Guntupalli SR. Treatment of epithelial ovarian cancer. BMJ. 2020;371:m3773.
Kandalaft LE, Odunsi K, Coukos G. Immunotherapy in ovarian cancer: Are We There Yet? J Clin Oncol. 2019;37:2460–71.
Kim H, Xu H, George E, Hallberg D, Kumar S, Jagannathan V, et al. Combining PARP with ATR inhibition overcomes PARP inhibitor and platinum resistance in ovarian cancer models. Nat Commun. 2020;11:3726.
Vergote I, Gonzalez-Martin A, Lorusso D, Gourley C, Mirza MR, Kurtz JE, et al. Clinical research in ovarian cancer: consensus recommendations from the Gynecologic Cancer InterGroup. Lancet Oncol. 2022;23:e374–e84.
Natanzon Y, Goode EL, Cunningham JM. Epigenetics in ovarian cancer. Semin Cancer Biol. 2018;51:160–9.
Chen B, Dragomir MP, Yang C, Li Q, Horst D, Calin GA. Targeting non-coding RNAs to overcome cancer therapy resistance. Signal Transduct Target Ther. 2022;7:121.
Liu SJ, Dang HX, Lim DA, Feng FY, Maher CA. Long noncoding RNAs in cancer metastasis. Nat Rev Cancer. 2021;21:446–60.
Guo W, Wang Y, Yang M, Wang Z, Wang Y, Chaurasia S, et al. LincRNA-immunity landscape analysis identifies EPIC1 as a regulator of tumor immune evasion and immunotherapy resistance. Sci Adv. 2021;7:eabb3555.
Lee S, Kopp F, Chang TC, Sataluri A, Chen B, Sivakumar S, et al. Noncoding RNA NORAD regulates genomic stability by sequestering PUMILIO proteins. Cell. 2016;164:69–80.
Zhao J, Du P, Cui P, Qin Y, Hu C, Wu J, et al. LncRNA PVT1 promotes angiogenesis via activating the STAT3/VEGFA axis in gastric cancer. Oncogene. 2018;37:4094–109.
Mitra R, Chen X, Greenawalt EJ, Maulik U, Jiang W, Zhao Z, et al. Decoding critical long non-coding RNA in ovarian cancer epithelial-to-mesenchymal transition. Nat Commun. 2017;8:1604.
Zhao L, Ji G, Le X, Wang C, Xu L, Feng M, et al. Long noncoding RNA LINC00092 acts in cancer-associated fibroblasts to drive glycolysis and progression of ovarian cancer. Cancer Res. 2017;77:1369–82.
Wu Y, Guo Q, Ju X, Hu Z, Xia L, Deng Y, et al. HNRNPH1-stabilized LINC00662 promotes ovarian cancer progression by activating the GRP78/p38 pathway. Oncogene. 2021;40:4770–82.
Zhao L, Jiang L, Zhang M, Zhang Q, Guan Q, Li Y, et al. NF-κB-activated SPRY4-IT1 promotes cancer cell metastasis by downregulating TCEB1 mRNA via Staufen1-mediated mRNA decay. Oncogene. 2021;40:4919–29.
Shiromoto Y, Sakurai M, Minakuchi M, Ariyoshi K, Nishikura K. ADAR1 RNA editing enzyme regulates R-loop formation and genome stability at telomeres in cancer cells. Nat Commun. 2021;12:1654.
Vohhodina J, Goehring LJ, Liu B, Kong Q, Botchkarev VV Jr, Huynh M, et al. BRCA1 binds TERRA RNA and suppresses R-Loop-based telomeric DNA damage. Nat Commun. 2021;12:3542.
Wells JP, White J, Stirling PC. R loops and their composite cancer connections. Trends Cancer. 2019;5:619–31.
Santos-Pereira JM, Herrero AB, García-Rubio ML, Marín A, Moreno S, Aguilera A. The Npl3 hnRNP prevents R-loop-mediated transcription-replication conflicts and genome instability. Genes Dev. 2013;27:2445–58.
Gratacós FM, Brewer G. The role of AUF1 in regulated mRNA decay. Wiley Interdiscip Rev RNA. 2010;1:457–73.
Alfano L, Caporaso A, Altieri A, Dell’Aquila M, Landi C, Bini L, et al. Depletion of the RNA binding protein HNRNPD impairs homologous recombination by inhibiting DNA-end resection and inducing R-loop accumulation. Nucleic Acids Res. 2019;47:4068–85.
Hua W, Zhao Y, Jin X, Yu D, He J, Xie D, et al. METTL3 promotes ovarian carcinoma growth and invasion through the regulation of AXL translation and epithelial to mesenchymal transition. Gynecol Oncol. 2018;151:356–65.
Au Yeung CL, Co NN, Tsuruga T, Yeung TL, Kwan SY, Leung CS, et al. Exosomal transfer of stroma-derived miR21 confers paclitaxel resistance in ovarian cancer cells through targeting APAF1. Nat Commun. 2016;7:11150.
Shibue T, Weinberg RA. EMT, CSCs, and drug resistance: the mechanistic link and clinical implications. Nat Rev Clin Oncol. 2017;14:611–29.
White EJ, Matsangos AE, Wilson GM. AUF1 regulation of coding and noncoding RNA. Wiley Interdiscip Rev RNA. 2017;8. https://doi.org/10.1002/wrna.1393.
Xin H, Brown JA, Gong C, Fan H, Brewer G, Gnarra JR. Association of the von Hippel-Lindau protein with AUF1 and posttranscriptional regulation of VEGFA mRNA. Mol Cancer Res. 2012;10:108–20.
Fu XD, Ares M Jr. Context-dependent control of alternative splicing by RNA-binding proteins. Nat Rev Genet. 2014;15:689–701.
Yoon JH, De S, Srikantan S, Abdelmohsen K, Grammatikakis I, Kim J, et al. PAR-CLIP analysis uncovers AUF1 impact on target RNA fate and genome integrity. Nat Commun. 2014;5:5248.
Crossley MP, Bocek M, Cimprich KA. R-Loops as cellular regulators and genomic threats. Mol Cell. 2019;73:398–411.
Marnef A, Legube G. R-loops as Janus-faced modulators of DNA repair. Nat Cell Biol. 2021;23:305–13.
Cerritelli SM, Crouch RJ. Ribonuclease H: the enzymes in eukaryotes. FEBS J. 2009;276:1494–505.
Ransohoff JD, Wei Y, Khavari PA. The functions and unique features of long intergenic non-coding RNA. Nat Rev Mol Cell Biol. 2018;19:143–57.
Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–307.
Wang X, Li J, Bian X, Wu C, Hua J, Chang S, et al. CircURI1 interacts with hnRNPM to inhibit metastasis by modulating alternative splicing in gastric cancer. Proc Natl Acad Sci USA. 2021;118:e2012881118.
Chen L, Rashid F, Shah A, Awan HM, Wu M, Liu A, et al. The isolation of an RNA aptamer targeting to p53 protein with single amino acid mutation. Proc Natl Acad Sci USA. 2015;112:10002–7.
Wang H, Fang L, Jiang J, Kuang Y, Wang B, Shang X, et al. The cisplatin-induced lncRNA PANDAR dictates the chemoresistance of ovarian cancer via regulating SFRS2-mediated p53 phosphorylation. Cell Death Dis. 2018;9:1103.
Li N, Zhan X, Zhan X. The lncRNA SNHG3 regulates energy metabolism of ovarian cancer by an analysis of mitochondrial proteomes. Gynecol Oncol. 2018;150:343–54.
Matsui M, Sakasai R, Abe M, Kimura Y, Kajita S, Torii W, et al. USP42 enhances homologous recombination repair by promoting R-loop resolution with a DNA-RNA helicase DHX9. Oncogenesis. 2020;9:60.
Sessa G, Gómez-González B, Silva S, Pérez-Calero C, Beaurepere R, Barroso S, et al. BRCA2 promotes DNA-RNA hybrid resolution by DDX5 helicase at DNA breaks to facilitate their repair. EMBO J. 2021;40:e106018.
Yu Z, Mersaoui SY, Guitton-Sert L, Coulombe Y, Song J, Masson JY, et al. DDX5 resolves R-loops at DNA double-strand breaks to promote DNA repair and avoid chromosomal deletions. NAR Cancer. 2020;2:zcaa028.
Brickner JR, Garzon JL, Cimprich KA. Walking a tightrope: The complex balancing act of R-loops in genome stability. Mol Cell. 2022;82:2267–97.
Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability-an evolving hallmark of cancer. Nat Rev Mol Cell Biol. 2010;11:220–8.
Britton S, Dernoncourt E, Delteil C, Froment C, Schiltz O, Salles B, et al. DNA damage triggers SAF-A and RNA biogenesis factors exclusion from chromatin coupled to R-loops removal. Nucleic Acids Res. 2014;42:9047–62.
Skourti-Stathaki K, Proudfoot NJ. A double-edged sword: R loops as threats to genome integrity and powerful regulators of gene expression. Genes Dev. 2014;28:1384–96.
Liao B, Hu Y, Brewer G. Competitive binding of AUF1 and TIAR to MYC mRNA controls its translation. Nat Struct Mol Biol. 2007;14:511–8.
Loflin P, Chen CY, Shyu AB. Unraveling a cytoplasmic role for hnRNP D in the in vivo mRNA destabilization directed by the AU-rich element. Genes Dev. 1999;13:1884–97.
Mehta M, Raguraman R, Ramesh R, Munshi A. RNA binding proteins (RBPs) and their role in DNA damage and radiation response in cancer. Adv Drug Deliv Rev. 2022;191:114569.
Huang J, Zhang A, Ho TT, Zhang Z, Zhou N, Ding X, et al. Linc-RoR promotes c-Myc expression through hnRNP I and AUF1. Nucleic Acids Res. 2016;44:3059–69.
Castellanos-Rubio A, Fernandez-Jimenez N, Kratchmarov R, Luo X, Bhagat G, Green PH, et al. A long noncoding RNA associated with susceptibility to celiac disease. Science. 2016;352:91–5.
Yokoi A, Villar-Prados A, Oliphint PA, Zhang J, Song X, De Hoff P, et al. Mechanisms of nuclear content loading to exosomes. Sci Adv. 2019;5:eaax8849.
Möller A, Lobb RJ. The evolving translational potential of small extracellular vesicles in cancer. Nat Rev Cancer. 2020;20:697–709.
Funding
This study is supported by the National Key Research and Development Program (2019YFA0802600 and 2022YFC2403400), the National Natural Science Foundation of China (82172773, 32270590, 31930019), the Anhui Provincial Key Research and Development Program (2022e07020013), Natural Science Foundation of Anhui Province (2208085QH252), the 2020 USTC Affiliated Hospital Introduction Project to Medical Leading Technology (2020LXJS-05), Beijing Science & Technology Innovation Fund (KC2021-JX-0186-143), Natural Science Research Project of Colleges and Universities in Anhui Province (2022AH051255).
Author information
Authors and Affiliations
Contributions
GS, LC and YZ conceived, designed, and supervised all the experiments. HYL, and SSD conducted the experiments and most of the analysis. XLY, YL, LLQ, and YYW analyzed the remaining analysis. TJZ collected clinical samples. HYL, LC wrote the original draft of the manuscript. HYL, GS, LC, and YZ revised the manuscript and provided critical discussions. GS, LC and YZ provided the fundings for this study. All authors read, discussed and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
YZ, LC and GS have an ownership interest in a patent related to this research.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liu, H., Deng, S., Yao, X. et al. Ascites exosomal lncRNA PLADE enhances platinum sensitivity by inducing R-loops in ovarian cancer. Oncogene 43, 714–728 (2024). https://doi.org/10.1038/s41388-024-02940-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-024-02940-6