Abstract
Recent studies have shown that major depressive disorder (MDD) is accompanied by alterations in functional and structural network gradients. However, whether changes are present in the cortical morphometric similarity (MS) network gradient, and the relationship between alterations of the gradient and gene expression remains largely unknown. In this study, the MS network was constructed, and its gradient was calculated in 71 patients with first-episode, treatment-naive MDD, and 69 demographically matched healthy controls. Between-group comparisons were performed to investigate abnormalities in the MS network gradient, and partial least squares regression analysis was conducted to explore the association between gene expression profiles and MS network gradient-based alternations in MDD. We found that the gradient was primarily significantly decreased in sensorimotor regions in patients with MDD compared with healthy controls, and increased in visual-related regions. In addition, the altered principal MS network gradient in the left postcentral cortex and right lingual cortex exhibited significant correlations with symptom severity. The abnormal gradient pattern was spatially correlated with the brain-wide expression of genes enriched for neurobiologically relevant pathways, downregulated in the MDD postmortem brain, and preferentially expressed in different cell types and cortical layers. These results demonstrated alterations of the principal MS network gradient in MDD and suggested the molecular mechanisms for structural alternations underlying MDD.
Similar content being viewed by others
Introduction
Major depressive disorder (MDD) is a prevalent and debilitating psychiatric disease worldwide that causes major social and economic burdens on individuals and countries [1]. Despite substantial efforts, the pathophysiological mechanism behind MDD is still unclear due to the inconsistency of abnormalities and treatment effects on brain structure and function [2, 3].
While heterogeneous, a large body of recent magnetic resonance imaging (MRI) studies have reported widespread abnormalities in brain structure and function in MDD [4,5,6,7,8]. More specifically, individuals with MDD showed abnormal gray matter volumes [9], cortical thickness [10], surface area [11], structural networks [12, 13], and functional networks [14, 15] compared to healthy controls. Although converging evidence points to disrupted structure and function in regions within the cortico-mesolimbic circuit, the neurobiological mechanisms responsible for the interaction of low-level sensory processing and high-order cognitive remains unclear [16, 17].
The hierarchical architecture of macroscopic brain features follows a “sensory-fugal” axis, which spans from sensory-motor to multimodal areas, allowing for information encoding and integration from sensation to cognition [18, 19]. This architecture is assumed to aid the integration of abstract concepts, cognition, and behavior [20], and revealing it offers the possibility to shed light on how the integrated nature of neural processing gives rise to function and dysfunction. More recently, the advent of the diffusion map embedding method has allowed macroscale brain features to be converted to low-dimensional manifold representations, namely, gradients [21] that represent hierarchical organizing axes to describe continuous spectra of brain features. To date, an increasing number of gradient studies have attempted to investigate spatial transitions of brain organization. Among them, functional connectivity gradients reflect the functional spectrum from perception and action to abstract cognition [22], and a previous study found alterations in functional connectivity gradients in MDD [23].
In parallel, structural connectivity gradients capture gradual transitions of morphology or microstructure, and have the potential to reveal mechanisms of neurodevelopment [22, 24]. Traditionally, structural connectivity gradients were calculated based on structural covariance [25] or anatomical connectivity [26, 27]. However, structural covariance based on a large sample size for accuracy neglects gradient construction at the individual-subject level [28], and anatomical connectivity constructed by tractography based on diffusion-weighted MRI (DWI) under-recovers long-distance projections [29]. Morphometric similarity (MS) has recently been proposed to quantify the similarity between cortical areas, by combining multiple individual morphological features instead of measuring the interregional correlation of a single MRI feature across individuals [30]. MS networks contained more connections between cortical areas within the same cytoarchitectonic class than structural covariance and anatomical connectivity. The connections of MS networks correlate highly with the brain-wide expression of genes enriched for neurobiologically relevant pathways, and regions of higher MS have been shown more likely to have axon trajectories with each other in a rhesus monkey model [30]. In addition, MS networks can be viewed as a neuroimaging phenotype linking brain structural changes to transcriptional data [31, 32]. A recent study found that the principal MS gradient is anchored by sensory and motor cortices, and it is closely associated with properties of cortical organization in healthy subjects [33]. Nevertheless, whether and how the principal MS gradient is disrupted in patients with MDD remains unclear.
Emerging evidence suggests that genes play an important role in human brain networks, especially in shaping functionally valuable and metabolically costly connections [34, 35]. The availability of the whole-brain atlas of gene expression based on the Allen Human Brain Atlas (AHBA) database has generated new opportunities to understand how disease-related gene expression at the microlevel is associated with macrolevel brain alterations in various psychiatric disorders [36,37,38]. In addition, neuroimaging features as intermediate phenotypes are theoretically closer to genetic substrates of MDD [39]. Thus, more recently there has been an increasing number of imaging transcriptomics studies linking cortical morphology and functional alterations in MDD to gene expression data [23, 40], in an attempt to reveal the complex mechanisms behind MDD.
In the present study, we hypothesized that there would be significant differences in the principal MS gradient between patients with MDD and healthy controls. We further hypothesized that these differences would be related to gene expression. To test these hypotheses, we first combined the MS network and the diffusion map embedding method to construct the principal MS gradient and evaluate the hierarchical organization of the MS gradient in patients with MDD and healthy controls. The relationships between case-control changes in the principal MS gradient and clinical variables of symptoms were also examined. Second, we conducted partial least squares (PLS) regression to link MDD-related changes in the principal MS gradient with anatomically patterned gene expression to determine which genes are MDD-related. Moreover, we performed various enrichment analyses connecting genes, molecular pathways, cell classes, and cortical layers to further interpret our results. An overview of the analytical framework is summarized in Fig. 1.
A Gradient construction. Morphological features (GM, SA, CT, IC, MC) were first derived from individual structural imaging maps. Based on the DK atlas, regionally morphological features were extracted and concatenated into a vector in each region. Pearson’s correlation was calculated between each pair of regional vectors, and the MS matrix was obtained from each subject. Then, the MS matrix was transformed into the affinity matrix by using a kernel function. Finally, the diffusion embedding algorithm was applied to decompose the affinity matrix and the first gradient map was acquired from each subject. B Gene expression. The expression value in each region (the left hemisphere only) of each gene was extracted from the AHBA database, and the gene expression matrix could be obtained. C Transcriptional analysis. PLS regression was conducted to link MDD abnormalities of the principal MS gradient with gene expression data, and enrichment analyses were performed subsequently on the significant gene list of the first component of PLS (PLS1). AHBA Allen Human Brain Atlas, CT cortical thickness, DK Desikan-Killiany, GM gray matter, IC intrinsic (Gaussian) curvature, MC mean curvature, MDD major depressive disorder, MS morphometric similarity, PLS partial least squares, SA surface area.
Methods and materials
Participants
This study was approved by the ethical committee of Tianjin Medical University General Hospital, and written informed consent was obtained from participants. Seventy-two first-episode, treatment-naive patients with MDD were recruited, and diagnoses were determined by trained psychiatrists using the diagnostic criteria of the Structured Clinical Interview of DSM-IV (SCID). Following a previous study [41], we recruited only MDD patients with the Hamilton Depression Rating Scale-17 (HDRS-17) scores ≥ 18. The Hamilton Anxiety Rating Scale-14 (HARS-14) was also applied to evaluate anxiety symptoms in these patients, but we did not set a cutoff value. Seventy age- and gender-matched healthy controls were recruited from nearby communities, and they were free of any history of neurological and psychiatric disorders. The general exclusion criteria were (i) younger than 18 years or older than 65 years; (ii) left-handedness; (iii) the presence of magnetic resonance contraindications; (iv) organic intracranial lesions; (v) poor image quality checked before and during neuroimaging data preprocessing, and (vi) any history of comorbid alcohol or drug abuse.
Neuroimaging data acquisition and preprocessing
The MRI data of all participants were collected using a 3.0-Tesla GE Discovery MR750 scanner. For T1-weighted imaging acquisition, please see Supplementary Text. The 3D high-resolution T1-weighted images were preprocessed for cortical surface reconstruction using FreeSurfer (v6.0), including skull stripping, tissue segmentation, surface reconstruction, metric reconstruction, and spherical normalization parameter estimation (Supplementary Text).
Construction of MS gradients
The cortical surfaces were divided into 1533 spatially contiguous regions [33, 42] derived from the DK-68 atlas [43], which parcellated approximately 1 cm2 for each region by using a backtracking algorithm [44]. This parcellated DK anatomical template was transformed from standard space to each participant’s individual space by using inverse spherical normalization parameters estimated in cortical surface reconstruction, and five structural features were extracted including gray matter (GM) volume, cortical thickness (CT), surface area (SA), intrinsic (Gaussian) curvature (IC), and mean curvature (MC). Each participant’s morphometric feature was z-score normalized across 1533 regions, and Pearson’s correlation coefficients were calculated between each pair of z-score normalized morphometric feature vectors, which formed a 1533 × 1533 MS matrix for each participant. The construction of MS gradients was preprocessed using the BrainSpace toolbox [42]. Given that the principal MS gradient is closely associated with cortical fundamental properties and diverges from the principal functional gradient [33], we focused on MDD-related alterations in the principal MS gradient. Thus, the group-level scores of the principal MS gradient were finally generated across aligned patients with MDD and healthy individuals (Supplementary Text).
The MS gradient comparison
A general linear model (GLM) was used to investigate between-group alterations of the regional principal MS gradient while regressing out the effects of age, sex, and age × sex. In addition, to contextualize the regional principal MS gradient alterations in the MDD group, we referred them to two prior classifications of cortical areas: the Yeo atlas of the cortex classified according to resting-state networks [45], and the von Economo atlas of the cortex classified by cytoarchitectonic criteria [46]. To this end, the mean principal MS gradient score of all regions within a particular Yeo network or von Economo class was calculated, and the GLM was applied to explore case-control differences in the principal MS gradient by regressing out the same covariates. The significance of each region, network, or class was corrected with the Benjamini–Hochberg false discovery rate (BH-FDR) method p < 0.05.
Correlations between the abnormal principal MS gradient and clinical variables
The mean scores of brain regions with the abnormal principal MS gradient in the MDD group were extracted to perform partial correlations between the principal MS gradient and clinical variables (the HDRS and the HARS) in patients with MDD while adjusting for age, sex, and age × sex. The correlation results were BH-FDR corrected at p < 0.05.
Gene expression data preprocessing
The available gene expression data of six postmortem brains with 3702 distinct samples were provided by the AHBA database [47]. We used the abagen toolbox (https://www.github.com/netneurolab/abagen) to process and map the transcriptomic data onto the 1533 parcellated brain regions [48]. Briefly, the preprocessing of gene expression data can be summarized as the follows: (i) update probe-to-gene annotations; (ii) intensity-based filter; (iii) probe selection; (iv) sample-to-region matching; (v) handling missing data; (vi) sample normalization; (vii) gene normalization; (viii) sample-to-region combination metric, and (ix) selection of stable genes. Finally, only 7443 genes remained (Supplementary Text). Since only two of the six brains of the AHBA database included samples from the right hemisphere, only the left hemisphere was considered (more details of six donors in the AHBA database are shown in Supplementary Table 7). Thus, a gene expression matrix of 767 regions × 7443 genes was finally used for further analyses.
Transcription-neuroimaging association analysis
We used PLS regression [49] to relate the gene expression of 7443 genes to case-control differences in the principal MS gradient (t values from 767 cortical regions in the left hemisphere). In the PLS regression model, the z-score normalized gene expression matrix (767 regions × 7443 genes) was taken as the independent variable, and the z-score normalized principal MS gradient case-control t vector (767 regions × 1) was treated as the dependent variable. The PLS components, the linear combination of weighted gene expression values, are ranked by the explained variances between independent and dependent variables. Thus, the first PLS component (PLS1) provides the optimal low-dimensional representation of the covariance of the high-dimensional data matrices [50]. We used a spatial autocorrelation-preserving permutation test (i.e., spin test, n = 10,000) to examine whether the explained variance of the PLS component was significantly greater than expected by chance [51]. Moreover, a bootstrapping method was conducted to evaluate the significance of genes contributing to components (Supplementary Text). Only significant genes (BH-FDR p < 0.01) were retained for subsequent analyses.
To further explore the relationships between the expression of MDD-related genes and principal MS gradient alterations, we obtained 24 genes related to depression from the AHBA database [52] (https://help.brain-map.org/download/attachments/2818165/HBA_ISH_GeneList.pdf?version=2&modificationDate=1614977648535&api=v2), and 270 genes from the two recent genome-wide association studies (GWAS) of MDD [53, 54]. The spatial relationships between the expression of these genes (genes overlapped with significant PLS genes) and the principal MS gradient case-control differences in the left hemisphere were examined. In addition, we performed gene set analyses to further test whether the significant genes with positive or negative weight (PLS + /−) were enriched for MDD-related genes from the AHBA database [52] and risk genes identified by the largest GWASMDD [54] (Supplementary Text).
Enrichment analyses
To test whether significant PLS + /− genes were enriched for genes implicated in the pathogenesis of MDD and other brain disorders, we used a catalog of genes with differential expression information (fold change values) for five major psychiatric disorders [55]. Based on the analysis results (please see RESULTS section), only genes with positive weights in the first PLS component (PLS1 + ) were used in the following enrichment analyses. Next, Gene Ontology (GO), Reactome, and the Kyoto Encyclopedia of Genes and Genomes (KEGG) embedded in Metascape (https://metascape.org/gp/index.html#/main/step1) were used for functional annotations of the PLS1 + genes [56]. Cell-type specific expression analysis of the PLS1 + gene list was performed using the specificity index probability (pSI) package in R [57]. Cortical layer enrichment was applied by using marker genes obtained based on a previous transcriptomic study [58]. To explore developmental time windows across brain regions, developmental gene expression enrichment analysis was performed by the cell-type specific expression analysis (CSEA, http://genetics.wustl.edu/jdlab/csea-tool-2/) [57]. All enrichment analyses were corrected by BH-FDR p < 0.05 (Supplementary Text).
Null model
A spin test was conducted to control the potential confounding effects of spatial autocorrelations [51, 59]. A set of null Pearson’s correlation coefficients can be generated by randomly rotating the spherical projection of spatial maps while preserving the spatial relationship. Thus, in this study, we first performed 10,000 spin test permutations of cortical regions to generate a null distribution, and the pspin value was computed as the proportion of null values of the correlation coefficient that were greater than the real values of the correlation coefficient.
Reproducibility analyses
To test the reliability of our results, the following five analyses were performed: (i) validation of the robustness of the case-control principal MS gradient after regressing the effect of total intracranial volume (TIV); (ii) validation of the robustness of MS network construction with Spearman’s correlation analysis; (iii) testing the reliability of the principal MS gradient using an independent neuroimaging dataset [33]; (iv) testing the reliability of the association between gene expression and the case-control t-map of the principal MS gradient using three MDD-related gene markers of somatostatin (SST) interneurons [40], and (v) testing the reliability of enrichment results using a multi-gene-list meta-analysis [56] based on the GWASMDD [53, 54], and gene-category enrichment analysis (GCEA) with ensemble-based null models [60] (Supplementary Text).
Results
Data samples
After qualifying for the structural MRI data, we ultimately recruited 71 patients with MDD and 69 healthy subjects in this study (one MDD and one healthy control were excluded due to the artifact of head motion). The demographic and clinical data of patients and controls are shown in Supplementary Table 1. The two groups did not show significant differences in sex (chi-squared test, χ2 = 0.23, p = 0.63) or age (two-sample t-test, t = −0.28, p = 0.78).
Case-control differences in the principal MS gradient
The first principal gradient explained 34% of the MS network variance in our dataset (with similar variance explained in MDD and controls across gradients, revealed by the two-sample t-test, t = 4.34 × 10−16, p > 0.99, Supplementary Fig. 1). It also showed two extreme ends by motor and sensory cortices with the association cortex in the middle (Fig. 2A). The spatial patterns of the 2–5 gradients are shown in Supplementary Fig. 2.
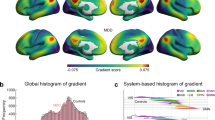
A The principal MS gradient pattern in patients with MDD and healthy controls. Motor and sensory cortices anchor at two extreme ends, with the association cortex in the middle. Regions with similar connectivity patterns show similar colors. B The histogram shows the distributions of mean principal MS gradient scores in the MDD and control group after regressing out the effect of age, sex, and age × sex. C Region-wise statistical comparisons between healthy controls and MDD, with healthy controls > MDD and healthy controls < MDD shown in red and blue, respectively (first row, un-threshold; second row, p < 0.05, threshold by BH-FDR correction). D A density scatterplot of the mean regional MS gradient scores of healthy controls (x-axis) and the case-control t-statistic (y-axis) (r = 0.46, pspin < 1 × 10−4). Most cortical regions have negative principal MS gradients in healthy controls, which decreases in healthy controls compared to MDD patients (34% of regions), or positive principal MS gradients in healthy controls, which increases in healthy controls compared to MDD patients (30% of regions). E Functional community-based absolute t-value (left, Yeo functional networks) and cytoarchitecture-based absolute t-value (right, von Economo classes) of the principal MS gradient indicate significant difference primarily in the somato-motor network and primary motor class. Asso1 association cortex1, Asso2 association cortex2, BH-FDR Benjamini–Hochberg false discovery rate, DAN dorsal attention network, DMN default mode network, FPN fronto-parietal network, Insula insular cortex, Limbic limbic regions, LN limbic network, MDD major depressive disorder, MS morphometric similarity, Prim motor primary motor cortex, Prim sens primary sensory cortex, Sec sens second sensory cortex, SMN somato-motor network, VAN ventral attention network, VN visual network.
Applying gradient alignment across individuals, we compared the principal MS gradient scores between patients with MDD and healthy controls using the GLM with age, sex, and age × sex as covariates. As shown in Fig. 2B, there was a significant case-control difference in the distribution of the mean principal MS gradient scores after regressing out age, sex, and age × sex (two-sample Kolmogorov–Smirnoff test, p = 1.11 × 10−4). Region-wise comparisons revealed that the principal MS gradient decreased in MDD in the left postcentral (part 15 and part 29), left frontal pole (part 2), right lateral occipital (part 43), and right precentral (part 25 and part 36) regions, and an increase in the principal MS gradient in MDD in the left pericalcarine (part 8), right cuneus (part 11), right lingual (part 17), right pericalcarine (part 7 and part 12), and right rostral middle frontal (part 24 and part 37) regions (Fig. 2C and Supplementary Table 2).
There was a positive correlation between the mean control regional MS gradient and the case-control t-map (r = 0.46, pspin < 1 × 10−4, Fig. 2D), which showed that regions with higher positive MS gradient scores in healthy controls tended to have a greater decrease in patients with MDD, and on the contrary, regions with higher negative MS gradient scores in healthy controls had a greater increase in MDD patients. This result indicated that higher MS gradient scores at the two extreme ends tend to show larger case-control differences.
We also applied two prior classifications of cortical regions (the Yeo 7 functional networks atlas [45] and the von Economo cytoarchitectural atlas [46]) to apply the findings to the functional and cytoarchitectural level of brain organization. For the Yeo functional networks atlas, patients with MDD had a decreased principal MS gradient in the somatomotor network (corrected p = 1.14 × 10−2, Fig. 2E and Supplementary Fig. 3 and Supplementary Table 3). For the von Economo cytoarchitectural atlas, patients with MDD exhibited a decreased principal MS gradient in the motor cytoarchitectural class (corrected p = 9.84 × 10−3, Fig. 2E and Supplementary Fig. 3 and Supplementary Table 4).
The correlations between case-control changes in the principal MS gradient scores and clinical variables were assessed by using partial correlation analysis controlling for age, sex, and age × sex. We included two clinical variables in this study: the HDRS-17 and the HARS-14. After BH-FDR correction, we found that the left postcentral cortex (part 15) exhibited a significant negative correlation with HDRS scores, whereas the right lingual cortex (part 17) had a significant positive correlation with HDRS scores (Supplementary Table 5). No significant correlations were found between the principal MS gradient scores and HARS scores.
Transcription-neuroimaging associations
The brain gene expression matrix was obtained from the AHBA database. Considering just two right hemisphere data available, we only used the left hemisphere in this study. Accordingly, the brain gene expression matrix (767 regions × 7443 genes) was applied to PLS regression to identify patterns of gene expression correlated with the anatomical distribution of the principal MS gradient of case-control differences (Fig. 3A). Only the first PLS component (PLS1) explained 8.5% of the variance in the principal MS gradient case-control differences, significantly more than expected by chance (spin test, pspin = 2.1 × 10−3). The distribution of the PLS1 scores showed an anterior-posterior gradient of gene expression (Fig. 3B), which is consistent with a previous study [31]. The PLS1 gene expression map was positively correlated with the case-control t-map (r = 0.29, pspin = 2.1 × 10−4, Fig. 3C). As suggested by previous studies [31, 36], we ranked the normalized weights of PLS1 according to the z score of each gene. Overall, 5485 genes were identified to make significant contributions to PLS1 (p < 0.01, BH-FDR corrected, Fig. 3D). Among them, 2882 genes had normalized positive PLS1 weights (PLS1 + ), and 2603 genes had normalized negative PLS1 weights (PLS1-), which indicated that gene expression was either over- or under-expressed, corresponding to increased or decreased changes in the principal MS gradient in the healthy control group compared to the MDD group.
A The case-control t-map of the regionally principal MS gradient scores in the left hemisphere. B A weighted gene expression map of regional PLS1 scores in the left hemisphere. C A density scatterplot shows the relationship between regional PLS1 scores and regional changes in the principal MS gradient (r = 0.29, pspin = 2 × 10−4). The gray band indicates the 95% confidence interval. D Ranked PLS1 genes based on z score. E Psychiatric enrichment analysis using the PLS1 + and PLS1- gene lists (the dashed line represents the significant threshold). AAD alcohol abuse or dependence, AHBA Allen Human Brain Atlas, ASD autism spectrum disorder, BD bipolar disorder, BH-FDR Benjamini–Hochberg false discovery rate, DGs differential genes, ISH in situ hybridization, MDD major depressive disorder, MS morphometric similarity, PLS partial least squares, SCZ schizophrenia.
Moreover, among 13 genes (the overlapped genes between MDD-related genes from the AHBA database and background genes), eight MDD-related genes from the AHBA database made significant contributions to PLS1 (Supplementary Table 6); among 221 genes (the overlapped genes between MDD-related genes from the GWASMDD and background genes), 92 MDD-related genes from the GWAS made significant contributions to PLS1. For MDD-related genes from the AHBA database, 46% (6 of 13) of the genes were in the PLS1 + gene list, and 15% (2 of 13) were in the PLS1- gene list. The expression profiles of all these genes (62%, 8 of 13) were significantly spatially correlated with the case-control t-map (all pspins < 0.05, BH-FDR corrected, Supplementary Fig. 4). For MDD-related genes from the GWAS, 19% (42 of 221) of the genes were in the PLS1 + gene list, and 23% (50 of 221) were in the PLS1- gene list. The expression profiles of part of these genes (39%, 86 of 221) were significantly spatially correlated with the case-control t-map (all pspins < 0.05, BH-FDR corrected, Supplementary Fig. 5). Besides, we found that there was no significant enrichment of the significant PLS1 genes for MDD-related genes from in situ hybridization (ISH) gene expression in the AHBA database (p = 0.90) or the PLS1- genes for risk genes identified by the GWASMDD (p = 0.94), but the PLS1 + genes were significantly enriched for risk genes identified by the GWASMDD (p = 0.029)
The PLS1 + gene list of the principal MS gradient denoted gene downregulation in patients with MDD
We first performed psychiatric disorder enrichment analysis to investigate whether the PLS1 + or PLS1- genes were enriched for genes with differential expression information for five major psychiatric disorders. As shown in Fig. 3E, the PLS1 + genes were significantly enriched for MDD downregulated genes (corrected p = 9.18 × 10−4), ASD upregulated genes (corrected p = 5.84 × 10−5), bipolar depression upregulated (corrected p = 1.96 × 10−2) and downregulated (corrected p = 5.08 × 10−6) genes, and schizophrenia upregulated genes (corrected p = 3.30 × 10−11). For the PLS1- genes, they were only enriched for genes with schizophrenia downregulated genes (corrected p = 1.06 × 10−2). Therefore, we only focus on the PLS1 + gene list for subsequent analysis based on the abovementioned results.
Pathway enrichment associated with changes in the principal MS gradient
Metascape was used for gene functional annotations, and the 15,633 genes with qualified brain expression data were used as background. The PLS1 + gene list was mainly enriched for several GO biological processes, Reactome gene sets, and KEGG pathways (Fig. 4A, B).
A The bubble plot shows the GO and KEGG functional annotations for the PLS1 + genes. The bubble size represents the number of overlapped genes between the PLS1 + gene list and each GO term, Reactome gene set, or KEGG pathway (y-axis). The color bar represents the BH-FDR corrected p value. B Metascape enrichment network visualization showing the intra-cluster and inter-cluster similarities of enriched pathways. Each pathway is shown by a node, where the node size is proportional to the number of input genes included in the pathway, and different colors respond to different clusters. *in A, B indicates that the biological processes are still significant by using ensemble-based null models in reproducibility analysis. C Cell-type specific expression analysis of the PLS1 + gene list. D Cortical layer enrichment analysis of the PLS1 + gene list. E Developmental gene expression enrichment analysis of the PLS1 + gene list. The bubble size is proportional to the BH-FDR corrected p value. The bubble color represents whether the PLS1 + genes are significantly enriched or not; the red represents yes, and the blue represents no. Ado adolescence, BH-FDR Benjamini–Hochberg false discovery rate, EC early childhood, EF early fetal, EMF early/mid fetal, F false, GO gene ontology, KEGG Kyoto Encyclopedia of Genes and Genomes, LF late fetal, LI late infancy, LMF late/mid fetal, M/LC mid/late childhood, NEF neonatal early infancy, PLS partial least squares, R-HAS Reactome-Homo sapiens, T true, YA young adulthood.
Cell-type specific expression associated with changes in the principal MS gradient
Given that the pathogenesis of MDD is complex and includes complicated interactions among diverse cell types in the brain, we used five central nervous system (CNS) cell types to further refine our analysis. Cell-type specific expression analysis revealed broad enrichment across astrocytes (corrected p = 2.12 × 10−51), neurons (corrected p = 8.23 × 10−18), and microglia (corrected p = 9.54 × 10−13, Fig. 4C).
Cortical layer enrichment associated with changes in the principal MS gradient
Using the laminar gene markers, we linked the PLS1 + genes with mesoscale cortical layers. The PLS1 + gene list was significantly enriched for layer I (corrected p = 7.18 × 10−16), layer II (corrected p = 3.48 × 10−7) and layer V (corrected p = 2.14 × 10−4, Fig. 4D).
Developmental gene expression enrichment associated with changes in the principal MS gradient
By using the web server CSEA, we investigated whether the PLS1 + genes were enriched in particular human brain regions or developmental windows. Developmental gene expression analysis revealed that the PLS1 + genes were expressed in the brain from early/mid fetal development onward and across several brain regions comprising the cortex and subcortex (i.e., thalamus, striatum, amygdala). The PLS1 + genes were expressed predominantly in late infancy and mid/late childhood (Fig. 4E).
Reproducibility analyses
To test the reliability of our results, we first examined whether case-control differences in the principal MS gradient were affected by TIV. As shown in Supplementary Fig. 6, the case-control differences in the principal MS gradient with TIV as a covariate were highly correlated to those without TIV (r = 0.99, pspin < 1 × 10−4).
Second, we conducted Spearman’s correlation analysis instead of Pearson’s correlation to construct the MS network and further validated the reliability of the case-control differences. The results showed that the case-control differences in the principal MS gradient based on Spearman’s correlation analysis were largely reproducible (r = 0.91, pspin < 1 × 10−4, Supplementary Fig. 7).
Third, we repeated the gradient construction using an independent neuroimaging dataset of healthy subjects provided by another study [33] (n = 116, mean age ± standard error of the mean (SEM) = 25.77 ± 0.61 years, 58 females) to test the reproducibility of the principal MS gradient. The results of the spatial correlation analysis indicated that the pattern of the principal MS gradient was reliable (r = 0.92, pspin < 1 × 10−4, Supplementary Fig. 8).
Fourth, we used the three canonical gene markers of SST interneurons (i.e., SST, CORT, NPY) to estimate the reliability of the association between gene expression and the case-control t-map of the principal MS gradient. The results showed that all three genes were contained in the PLS1 + gene list, and they were significantly correlated with the case-control t-map of the principal MS gradient (SST: r = 0.21, pspin = 2.3 × 10−2; NPY: r = 0.25, pspin = 1.3 × 10−2; CORT: r = 0.16, pspin = 1.7 × 10−2, all BH-FDR corrected, Supplementary Fig. 9).
Finally, we performed two validation analyses to further validate the reliability of gene enrichment results. For results of the multi-gene-list meta-analysis, we found that all GO categories, Reactome gene sets, and KEGG pathways of genes from GWASMDD overlapped with genes from the PLS1 + gene list (Supplementary Text and Supplementary Fig. 10). For results of GCEA using ensemble-based null models, 7/14 GO categories related to biological processes were still significant by using the more rigorous null models, which indicated the results of gene enrichment were relatively reliable (Supplementary Text, Fig. 4A, B, Supplementary Fig. 11, and Supplementary Tables 8 and 9).
Discussion
Here, we for the first time investigated the abnormalities of the principal MS gradient in first-episode, treatment-naive patients with MDD and healthy controls. We determined that the principal MS gradient map of case-control difference was spatially correlated to the cortical gene expression map. Furthermore, we found that genes related to MS gradient changes (PLS1 + ) were not only significantly enriched for downregulated genes in postmortem brain with MDD and neurobiologically relevant pathways, but also preferentially expressed in certainly different cell types and cortical layers, which offers a perspective on the relationship between the hierarchical organization of the macroscopically morphometric profile and the microscopic transcriptomes during the onset and progression of MDD.
One of the advantages of MS is the combination of structural multi-features in a single participant compared with structural covariance and anatomical connectivity [30, 32, 61]. The principal MS gradient pattern is closely associated with cortical fundamental properties, from gene expression, cytoarchitecture, and myeloarchitecture to evolutionary expansion [33]. Given that MDD is a disorder that disrupts neuroplasticity combined with multi-factors (e.g., genetics, gene-environmental interactions, neuroendocrine, and inflammation) [62], we used a complex neuroimaging phenotype, the principal MS gradient, to investigate differences between patients with MDD and healthy controls. Our principal MS gradient mapping showed that both groups revealed an axis of connectivity variations with motor and sensory cortices on two extreme ends and the association cortex in the middle (Fig. 2A). We found that alterations of the principal MS gradient were mainly concentrated in areas related to the somatomotor network (SMN), such as precentral and postcentral regions. One of the famous hallmarks of MDD is psychomotor abnormalities that designate the interaction of affective and cognitive functions with motor function [63, 64]. A series of previous studies suggested that neuronal activity in the motor cortex and SMN is closely associated with subcortical inputs and other nonmotor cortical networks, where the default mode network stands in the reciprocal position [64]. The alterations of the principal MS gradient in SMN again reiterate the core role of lower hierarchy regions involved in feedforward communication in the process of MDD development. Furthermore, abnormalities were also observed in the frontal pole and visual-related regions, which suggested that depression could cause disturbances in sensory perception systems, and disruptions in visual functions may serve as an essential clinical feature underlying MDD [65, 66]. In the present work, our findings of abnormalities in the frontal pole and visual network-related regions thought to be a part of low-level sensory perception systems may indicate that hierarchical organization changes in sensory systems could be endophenotypes in MDD pathology.
MDD emerges from the complex interactions of biological systems that span genes and molecules through cells, networks, and behaviors [40, 62, 67]. Thus, MDD-related alterations in the principal MS gradient might also be multiscale changes. By using the PLS approach, we found that the cortical map of case-control differences in the principal MS gradient was spatially correlated with the cortical gene expression map and further identified the weighted combination of genes in the first PLS component that may drive structural hierarchical organization changes mediating the genetic risk of MDD. Our results showed that the PLS1 + gene list was not only significantly enriched for downregulated genes in postmortem individuals with MDD but also significantly enriched for SCZ-, ASD-, and BD-dysregulated genes, which is in line with the results of a prior GWAS showing that psychiatric disorders share common variant risks [68, 69]. Furthermore, gene markers of SST interneurons were consistently spatially correlated with the changes in the principal MS gradient. Converging evidence suggests that SST interneurons are preferentially vulnerable in patients with depression and are the pronounced pathophysiological feature of depression [70, 71], which was further corroborated by our results. All of the abovementioned results indicate that downregulated genes of the postmortem brain in MDD were normally overexpressed in cortical areas with decreased principal MS gradient scores.
The genes in the PLS1 + list were highly expressed in the frontal and temporal cortical areas, which were significantly enriched for several relevant categories. The identified categories were mainly implicated in neuron development, axon guidance, and response to stimuli, indicating that neuronal impairment and maldevelopment may be possible mechanisms of MDD. In addition, we found that the significant PLS1 genes were not significantly enriched for MDD-related genes, which is possibly due to the limited number of MDD-related genes identified in the AHBA database. The significant enrichment of PLS1 + genes for the risk genes identified by the large-scale GWASMDD, along with several neurobiological categories shared between our study and the results of GWASMDD, highlights that the MDD-related genes identified by PLS may provide novel insight into the complex substrates of MDD.
Converging evidence indicates that understanding alterations in cortical neuronal positioning and the complex interplay of brain cells with specific contributions to psychiatric disease offers opportunities to identify new avenues for disease treatment [72, 73]. Using single-cell expression data and cortical layer markers, we observed that significant enrichment of MDD-related genes was most pronounced in astrocytes and neurons. Astrocytes have a close relationship with synaptic activity and modulate neuronal circuits and behavior, which play a key role in MDD [74]. Combining the results of pathway and cell enrichment, neuroinflammation and microglia were also identified, which underlines the mechanisms of inflammation relevant to the development of MDD [62]. We found that MDD-related genes were mainly enriched for layer I, layer II, and layer V, indicating that the characteristic distribution of different neurons and their connections with other regions may provide insight into the nature of the pathology in MDD [72, 73]. Moreover, developmental enrichment analyses revealed gene sets that were expressed in the cortex and several subcortical regions from the early/mid fetal stage to young adulthood in MDD by associating gene expression patterns with our macroscale findings, suggesting that potential time windows for susceptibility to MDD might be earlier than young adulthood.
Several limitations of this study should be highlighted. The first limitation is that we only used five morphometric features to construct the MS gradient. Although accumulating evidence reveals that T1 weighted-restricted MS network construction could be an appropriate proxy for multimodal MS networks when multimodal imaging is not available [32, 33, 75], it is still necessary for future studies to investigate the hierarchical organization of morphology with multimodal imaging data. Second, the sample size in the current study was relatively small, and future studies with large sample sizes are needed to further validate MS gradients across individuals and to identify abnormalities in patients with MDD. Third, since gene expression profiles are dependent on age, sex, and ethnicity [76, 77], these factors might bias the results of our study. Finally, we only validated the results of GO enrichment analysis with ensemble-based null models because the toolbox only supports GCEA with GO terms [60].
In summary, the results of the current study supported our hypotheses that alterations of the principal MS gradient existed in individuals with MDD, and these alterations were spatially related to gene expression. We further showed that MDD-related genes were enriched for neurobiologically relevant pathways, downregulated genes in MDD, and were preferentially expressed in different cell types and cortical layers. Together, our findings provide new insight into understanding the altered coordination of structure in patients with MDD and may offer a new endophenotype to further investigate the complex substrate of depression.
Data availability
Human gene expression data that supports the findings of this study are available in the Allen Human Brain Atlas database (http://human.brain-map.org/static/download). Dysregulated genes in the postmortem brain of messenger RNA are from the raw Gandal et al. [55] dataset (https://www.science.org/doi/suppl/10.1126/science.aad6469/suppl_file/aad6469_gandal_sm_data-table-s1.xlsx). Raw data of the expression data from different cell types was gained from GSE73721 (Gene Expression Omnibus, GEO). Different layer markers were from the raw He et al. [58] dataset (https://static-content.springer.com/esm/art%3A10.1038%2Fnn.4548/MediaObjects/41593_2017_BFnn4548_MOESM255_ESM.xlsx). The neuroimaging preprocessing software is freely available (FreeSurfer v6.0, http://surfer.nmr.mgh.harvard.edu). The codes for MS analysis and PLS are openly available at https://github.com/SarahMorgan/Morphometric_Similarity_SZ. The codes for the construction of MS gradients are available at https://github.com/MICA-MNI/BrainSpace. The codes for gene expression analysis can be found at https://github.com/rmarkello/abagen. The codes for gene set analysis using MAGMA software are available at https://github.com/SarahMorgan/Morphometric_Similarity_SZ/blob/master/magma_enrichments.md. Pathway enrichment analysis was analyzed at https://metascape.org. Cell-type-specific expression analysis (CSEA) was analyzed at http://genetics.wustl.edu/jdlab/csea-tool-2. The code for spatial permutation testing can be found at https://github.com/frantisekvasa/rotate_parcellation. The codes for performing GCEA are available at https://github.com/benfulcher/GeneCategoryEnrichmentAnalysis.
References
World Health O. The global burden of disease: 2004 update: World Health Organization; 2008.
Zhuo C, Li G, Lin X, Jiang D, Xu Y, Tian H, et al. The rise and fall of MRI studies in major depressive disorder. Transl Psychiatry. 2019;9:335.
Cipriani A, Furukawa TA, Salanti G, Chaimani A, Atkinson LZ, Ogawa Y, et al. Comparative efficacy and acceptability of 21 antidepressant drugs for the acute treatment of adults with major depressive disorder: a systematic review and network meta-analysis. Lancet. 2018;391:1357–66.
Fitzgerald PB, Laird AR, Maller J, Daskalakis ZJ. A meta-analytic study of changes in brain activation in depression. Hum Brain Mapp. 2008;29:683–95.
Geng J, Yan R, Shi J, Chen Y, Mo Z, Shao J, et al. Altered regional homogeneity in patients with somatic depression: A resting-state fMRI study. J Affect Disord. 2019;246:498–505.
Guo WB, Liu F, Xue ZM, Yu Y, Ma CQ, Tan CL, et al. Abnormal neural activities in first-episode, treatment-naïve, short-illness-duration, and treatment-response patients with major depressive disorder: a resting-state fMRI study. J Affect Disord. 2011;135:326–31.
Xue K, Liang S, Yang B, Zhu D, Xie Y, Qin W, et al. Local dynamic spontaneous brain activity changes in first-episode, treatment-naïve patients with major depressive disorder and their associated gene expression profiles. Psychol Med. 2022;52:2052–61.
Zhang FF, Peng W, Sweeney JA, Jia ZY, Gong QY. Brain structure alterations in depression: Psychoradiological evidence. CNS Neurosci Ther. 2018;24:994–1003.
Wise T, Radua J, Via E, Cardoner N, Abe O, Adams TM, et al. Common and distinct patterns of grey-matter volume alteration in major depression and bipolar disorder: evidence from voxel-based meta-analysis. Mol Psychiatry. 2017;22:1455–63.
Patel Y, Parker N, Shin J, Howard D, French L, Thomopoulos SI, et al. Virtual histology of cortical thickness and shared neurobiology in 6 psychiatric disorders. JAMA Psychiatry. 2021;78:47–63.
Schmaal L, Hibar DP, Sämann PG, Hall GB, Baune BT, Jahanshad N, et al. Cortical abnormalities in adults and adolescents with major depression based on brain scans from 20 cohorts worldwide in the ENIGMA Major Depressive Disorder Working Group. Mol Psychiatry. 2017;22:900–9.
Korgaonkar MS, Fornito A, Williams LM, Grieve SM. Abnormal structural networks characterize major depressive disorder: a connectome analysis. Biol Psychiatry. 2014;76:567–74.
Bai F, Shu N, Yuan Y, Shi Y, Yu H, Wu D, et al. Topologically convergent and divergent structural connectivity patterns between patients with remitted geriatric depression and amnestic mild cognitive impairment. J Neurosci. 2012;32:4307–18.
Qiu L, Xia M, Cheng B, Yuan L, Kuang W, Bi F, et al. Abnormal dynamic functional connectivity of amygdalar subregions in untreated patients with first-episode major depressive disorder. J Psychiatry Neurosci. 2018;43:262–72.
Yan CG, Chen X, Li L, Castellanos FX, Bai TJ, Bo QJ, et al. Reduced default mode network functional connectivity in patients with recurrent major depressive disorder. Proc Natl Acad Sci USA. 2019;116:9078–83.
Price RB, Duman R. Neuroplasticity in cognitive and psychological mechanisms of depression: an integrative model. Mol Psychiatry. 2020;25:530–43.
Miller CH, Hamilton JP, Sacchet MD, Gotlib IH. Meta-analysis of functional neuroimaging of major depressive disorder in youth. JAMA Psychiatry. 2015;72:1045–53.
Mesulam MM. From sensation to cognition. Brain. 1998;121:1013–52.
Mesulam M. Patterns in behavioral neuroanatomy: association areas, the limbic system, and hemispheric specialization. In: Mesulam M, editor. Principles of Behavioral Neurology. Philadelphia: F.A. Davis; 1985. p. 1–70.
Mesulam M. The evolving landscape of human cortical connectivity: facts and inferences. Neuroimage. 2012;62:2182–9.
Margulies DS, Ghosh SS, Goulas A, Falkiewicz M, Huntenburg JM, Langs G, et al. Situating the default-mode network along a principal gradient of macroscale cortical organization. Proc Natl Acad Sci USA. 2016;113:12574–9.
Huntenburg JM, Bazin PL, Margulies DS. Large-scale gradients in human cortical organization. Trends Cogn Sci. 2018;22:21–31.
Xia M, Liu J, Mechelli A, Sun X, Ma Q, Wang X, et al. Connectome gradient dysfunction in major depression and its association with gene expression profiles and treatment outcomes. Mol Psychiatry. 2022;27:1384–93.
Burt JB, Demirtaş M, Eckner WJ, Navejar NM, Ji JL, Martin WJ, et al. Hierarchy of transcriptomic specialization across human cortex captured by structural neuroimaging topography. Nat Neurosci. 2018;21:1251–9.
Valk SL, Xu T, Margulies DS, Masouleh SK, Paquola C, Goulas A, et al. Shaping brain structure: Genetic and phylogenetic axes of macroscale organization of cortical thickness. Sci Adv. 2020;6:eabb3417.
Paquola C, Seidlitz J, Benkarim O, Royer J, Klimes P, Bethlehem RAI, et al. A multi-scale cortical wiring space links cellular architecture and functional dynamics in the human brain. PLoS Biol. 2020;18:e3000979.
Park BY, Hong SJ, Valk SL, Paquola C, Benkarim O, Bethlehem RAI, et al. Differences in subcortico-cortical interactions identified from connectome and microcircuit models in autism. Nat Commun. 2021;12:2225.
Alexander-Bloch A, Raznahan A, Bullmore E, Giedd J. The convergence of maturational change and structural covariance in human cortical networks. J Neurosci. 2013;33:2889–99.
Dauguet J, Peled S, Berezovskii V, Delzescaux T, Warfield SK, Born R, et al. Comparison of fiber tracts derived from in-vivo DTI tractography with 3D histological neural tract tracer reconstruction on a macaque brain. Neuroimage. 2007;37:530–8.
Seidlitz J, Váša F, Shinn M, Romero-Garcia R, Whitaker KJ, Vértes PE, et al. Morphometric similarity networks detect microscale cortical organization and predict inter-individual cognitive variation. Neuron. 2018;97:231–47.
Li J, Seidlitz J, Suckling J, Fan F, Ji GJ, Meng Y, et al. Cortical structural differences in major depressive disorder correlate with cell type-specific transcriptional signatures. Nat Commun. 2021;12:1647.
Seidlitz J, Nadig A, Liu S, Bethlehem RAI, Vértes PE, Morgan SE, et al. Transcriptomic and cellular decoding of regional brain vulnerability to neurogenetic disorders. Nat Commun. 2020;11:3358.
Yang S, Wagstyl K, Meng Y, Zhao X, Li J, Zhong P, et al. Cortical patterning of morphometric similarity gradient reveals diverged hierarchical organization in sensory-motor cortices. Cell Rep. 2021;36:109582.
Thompson PM, Cannon TD, Narr KL, van Erp T, Poutanen VP, Huttunen M, et al. Genetic influences on brain structure. Nat Neurosci. 2001;4:1253–8.
Arnatkeviciute A, Fulcher BD, Oldham S, Tiego J, Paquola C, Gerring Z, et al. Genetic influences on hub connectivity of the human connectome. Nat Commun. 2021;12:4237.
Morgan SE, Seidlitz J, Whitaker KJ, Romero-Garcia R, Clifton NE, Scarpazza C, et al. Cortical patterning of abnormal morphometric similarity in psychosis is associated with brain expression of schizophrenia-related genes. Proc Natl Acad Sci USA. 2019;116:9604–9.
Xie Y, Zhang X, Liu F, Qin W, Fu J, Xue K, et al. Brain mRNA expression associated with cortical volume alterations in autism spectrum disorder. Cell Rep. 2020;32:108137.
Ji Y, Zhang X, Wang Z, Qin W, Liu H, Xue K, et al. Genes associated with gray matter volume alterations in schizophrenia. Neuroimage. 2021;225:117526.
Gottesman II, Gould TD. The endophenotype concept in psychiatry: etymology and strategic intentions. Am J Psychiatry. 2003;160:636–45.
Anderson KM, Collins MA, Kong R, Fang K, Li J, He T, et al. Convergent molecular, cellular, and cortical neuroimaging signatures of major depressive disorder. Proc Natl Acad Sci USA. 2020;117:25138–49.
Khan A, Bhat A, Kolts R, Thase ME, Brown W. Why has the antidepressant-placebo difference in antidepressant clinical trials diminished over the past three decades? CNS Neurosci Ther. 2010;16:217–26.
Vos de Wael R, Benkarim O, Paquola C, Lariviere S, Royer J, Tavakol S, et al. BrainSpace: a toolbox for the analysis of macroscale gradients in neuroimaging and connectomics datasets. Commun Biol. 2020;3:103.
Desikan RS, Ségonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage. 2006;31:968–80.
Romero-Garcia R, Atienza M, Clemmensen LH, Cantero JL. Effects of network resolution on topological properties of human neocortex. Neuroimage 2012;59:3522–32.
Yeo BT, Krienen FM, Sepulcre J, Sabuncu MR, Lashkari D, Hollinshead M, et al. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J Neurophysiol. 2011;106:1125–65.
Von Economo C, Koskinas GN. Atlas of cytoarchitectonics of the adult human cerebral cortex. 1st ed. Basel, Switzerland: Karger; 2008.
Hawrylycz MJ, Lein ES, Guillozet-Bongaarts AL, Shen EH, Ng L, Miller JA, et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012;489:391–9.
Markello RD, Arnatkeviciute A, Poline J-B, Fulcher BD, Fornito A, Misic B. Standardizing workflows in imaging transcriptomics with the abagen toolbox. eLife. 2021;10:e72129.
Abdi H. Partial least squares regression and projection on latent structure regression (PLS Regression). Wiley Interdiscip Rev: Computational Stat. 2010;2:97–106.
Abdi H, Williams LJ. Partial least squares methods: partial least squares correlation and partial least square regression. Methods Mol Biol. 2013;930:549–79.
Váša F, Seidlitz J, Romero-Garcia R, Whitaker KJ, Rosenthal G, Vértes PE, et al. Adolescent tuning of association cortex in human structural brain networks. Cereb Cortex. 2018;28:281–94.
Zeng H, Shen EH, Hohmann JG, Oh SW, Bernard A, Royall JJ, et al. Large-scale cellular-resolution gene profiling in human neocortex reveals species-specific molecular signatures. Cell. 2012;149:483–96.
Wray NR, Ripke S, Mattheisen M, Trzaskowski M, Byrne EM, Abdellaoui A, et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat Genet. 2018;50:668–81.
Howard DM, Adams MJ, Clarke TK, Hafferty JD, Gibson J, Shirali M, et al. Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat Neurosci. 2019;22:343–52.
Gandal MJ, Haney JR, Parikshak NN, Leppa V, Ramaswami G, Hartl C, et al. Shared molecular neuropathology across major psychiatric disorders parallels polygenic overlap. Science. 2018;359:693–7.
Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10:1523.
Dougherty JD, Schmidt EF, Nakajima M, Heintz N. Analytical approaches to RNA profiling data for the identification of genes enriched in specific cells. Nucleic Acids Res. 2010;38:4218–30.
He Z, Han D, Efimova O, Guijarro P, Yu Q, Oleksiak A, et al. Comprehensive transcriptome analysis of neocortical layers in humans, chimpanzees and macaques. Nat Neurosci. 2017;20:886–95.
Alexander-Bloch AF, Shou H, Liu S, Satterthwaite TD, Glahn DC, Shinohara RT, et al. On testing for spatial correspondence between maps of human brain structure and function. Neuroimage. 2018;178:540–51.
Fulcher BD, Arnatkeviciute A, Fornito A. Overcoming false-positive gene-category enrichment in the analysis of spatially resolved transcriptomic brain atlas data. Nat Commun. 2021;12:2669.
Wei Y, Scholtens LH, Turk E, van den Heuvel MP. Multiscale examination of cytoarchitectonic similarity and human brain connectivity. Netw Neurosci. 2019;3:124–37.
Otte C, Gold SM, Penninx BW, Pariante CM, Etkin A, Fava M, et al. Major depressive disorder. Nat Rev Dis Prim. 2016;2:1–20.
Yin Y, Wang M, Wang Z, Xie C, Zhang H, Zhang H, et al. Decreased cerebral blood flow in the primary motor cortex in major depressive disorder with psychomotor retardation. Prog Neuropsychopharmacol Biol Psychiatry. 2018;81:438–44.
Northoff G, Hirjak D, Wolf RC, Magioncalda P, Martino M. All roads lead to the motor cortex: psychomotor mechanisms and their biochemical modulation in psychiatric disorders. Mol Psychiatry. 2021;26:92–102.
Yang H, Chen X, Chen ZB, Li L, Li XY, Castellanos FX, et al. Disrupted intrinsic functional brain topology in patients with major depressive disorder. Mol Psychiatry. 2021;26:7363–71.
Lu F, Cui Q, Huang X, Li L, Duan X, Chen H, et al. Anomalous intrinsic connectivity within and between visual and auditory networks in major depressive disorder. Prog Neuropsychopharmacol Biol Psychiatry. 2020;100:109889.
Holmes AJ, Patrick LM. The myth of optimality in clinical neuroscience. Trends Cogn Sci. 2018;22:241–57.
Anttila V, Bulik-Sullivan B, Finucane HK, Walters RK, Bras J, Duncan L, et al. Analysis of shared heritability in common disorders of the brain. Science. 2018;360:eaap8757.
Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013;381:1371–9.
Fee C, Banasr M, Sibille E. Somatostatin-positive gamma-aminobutyric acid interneuron deficits in depression: cortical microcircuit and therapeutic perspectives. Biol Psychiatry. 2017;82:549–59.
Duman RS, Sanacora G, Krystal JH. Altered connectivity in depression: GABA and glutamate neurotransmitter deficits and reversal by novel treatments. Neuron. 2019;102:75–90.
Nagy C, Maitra M, Tanti A, Suderman M, Théroux JF, Davoli MA, et al. Single-nucleus transcriptomics of the prefrontal cortex in major depressive disorder implicates oligodendrocyte precursor cells and excitatory neurons. Nat Neurosci. 2020;23:771–81.
Chana G, Landau S, Beasley C, Everall IP, Cotter D. Two-dimensional assessment of cytoarchitecture in the anterior cingulate cortex in major depressive disorder, bipolar disorder, and schizophrenia: evidence for decreased neuronal somal size and increased neuronal density. Biol Psychiatry. 2003;53:1086–98.
Zhou X, Xiao Q, Xie L, Yang F, Wang L, Tu J. Astrocyte, a promising target for mood disorder interventions. Front Mol Neurosci. 2019;12:136.
King DJ, Wood AG. Clinically feasible brain morphometric similarity network construction approaches with restricted magnetic resonance imaging acquisitions. Netw Neurosci. 2020;4:274–91.
Mogil LS, Andaleon A, Badalamenti A, Dickinson SP, Guo X, Rotter JI, et al. Genetic architecture of gene expression traits across diverse populations. PLoS Genet. 2018;14:e1007586.
Somel M, Guo S, Fu N, Yan Z, Hu HY, Xu Y, et al. MicroRNA, mRNA, and protein expression link development and aging in human and macaque brain. Genome Res. 2010;20:1207–18.
Acknowledgements
We are grateful to all the participants for their cooperation and patience. We also thank Wei Liao and Siqi Yang (University of Electronic Science and Technology of China) for the support of data providing and graphic presentation.
Funding
This work was funded by Natural Science Foundation of China (82072001 and 82001796) and Tianjin Key Medical Discipline (Specialty) Construction Project (TJYXZDXK-001A). The authors have nothing to disclose.
Author information
Authors and Affiliations
Contributions
KX, YZ, and FL conceptualized the study. KX conducted the analysis, and wrote the first version of the manuscript. LG and WZ helped analyze data and revise the manuscript. SL, QX, ML, and LM collected imaging data. YZ and FL provided critical comments on the manuscript and approved the final version of the manuscript. All authors contributed to the manuscript. The authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Xue, K., Guo, L., Zhu, W. et al. Transcriptional signatures of the cortical morphometric similarity network gradient in first-episode, treatment-naive major depressive disorder. Neuropsychopharmacol. 48, 518–528 (2023). https://doi.org/10.1038/s41386-022-01474-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41386-022-01474-3
This article is cited by
-
Exploring functional dysconnectivity in schizophrenia: alterations in eigenvector centrality mapping and insights into related genes from transcriptional profiles
Schizophrenia (2024)
-
Transcriptional signatures of the whole-brain voxel-wise resting-state functional network centrality alterations in schizophrenia
Schizophrenia (2023)
-
Integrating genetics and transcriptomics to study major depressive disorder: a conceptual framework, bioinformatic approaches, and recent findings
Translational Psychiatry (2023)