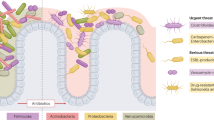
Abstract
The diverse bacterial communities that colonize the gastrointestinal tract play an essential role in maintaining immune homeostasis through the production of critical metabolites such as short-chain fatty acids (SCFAs) and this can be disrupted by antibiotic use. However, few studies have addressed the effects of specific antibiotics longitudinally on the microbiome and immunity. We evaluated the effects of four specific antibiotics: enrofloxacin, cephalexin, paromomycin, and clindamycin, in healthy female rhesus macaques. All antibiotics disrupted the microbiome, including reduced abundances of fermentative bacteria and increased abundances of potentially pathogenic bacteria, including Enterobacteriaceae in the stool, and decreased Helicobacteraceae in the colon. This was associated with decreased SCFAs, indicating altered bacterial metabolism. Importantly, antibiotic use also substantially altered local immune responses, including increased neutrophils and Th17 cells in the colon. Furthermore, we observed increased soluble CD14 in plasma, indicating microbial translocation. These data provide a longitudinal evaluation of antibiotic-induced changes to the composition and function of colonic bacterial communities associated with specific alterations in mucosal and systemic immunity.
Similar content being viewed by others
Introduction
The mammalian gastrointestinal (GI) tract is densely colonized with diverse bacterial communities, which are critical in human health, and contribute to the maintenance of mucosal and systemic immune homeostasis.1 Furthermore, compounds derived from the GI microbiota play key roles in promoting proper host immune function. Short-chain fatty acids (SCFAs), produced exclusively through microbial fermentation of dietary fibers in the colon, act through a variety of mechanisms to promote mucosal health. For example, butyrate is a key energy source for colonic epithelial cells and also influences gene expression by acting as a histone deacetylase inhibitor.2
In many disease states, there is an observed alteration to the abundance and diversity of the bacterial species in these communities when compared with healthy controls. One example is that of human immunodeficiency virus (HIV) infection, which has been associated with increased abundances of Proteobacteria and reduced abudances of Bacteroides. Aside from changes in bacterial community composition, HIV infection is associated with altered host–microbiota interactions. Specifically, impaired mucosal immunity and breaches in the epithelium lead to translocation of microbes and microbial products (microbial translocation, MT) from the colonic lumen into systemic circulation, which is associated with aberrant immune activation, and increased morbidity and mortality.3,4,5,6,7
Nonhuman primates (NHPs) are an extremely important model for critical research pertaining to human health, such as aging, viral hepatitis infection, Zika virus infection, and HIV infection.8,9,10 Indeed, Simian immunodeficiency virus (SIV) infection of macaques has been essential for understanding the pathogenesis of HIV infection. As in humans, NHP demonstrate colonic microbiota dysbiosis and MT during SIV infection,11,12,13,14,15 which has provided a detailed understanding of the consequences of MT, such as liver damage resulting from increased immune activation in response to increased burden of microbial products.16 SIV infection of NHP has also been critical for evaluating vaccines and therapeutic approaches.17 As more NHP studies are beginning to incorporate microbiome analyses, it is important to develop a detailed understanding of the composition of NHP microbiota and the effects of xenobiotics on these communities.
Antibiotics are important xenobiotics that are widely used in NHP veterinary and human healthcare, but are known to disrupt native microbiota.18 Due to the extensive interaction of the microbiota with the host immune system, these antibiotic-mediated alterations to the microbiota composition can result in distinct immunological consequences. For example, antibiotic treatment in mice resulted in increased inflammation as a result of increased MT and increased mucosal immune activation in the colon.19,20 Other murine models have demonstrated that administration of antibiotics early in life can increase incidence of autoimmune disorders.21 Indeed, our group previously demonstrated that antibiotics altered mucosal bacterial communities in SIV-infected rhesus macaques (RMs).17,22 However, due to the SIV infection, we were unable to attribute any immunological changes to the antibiotics alone. Thus, the impact of antibiotic treatments on the microbiota and mucosal immunity in NHP models has not been thoroughly evaluated, despite the importance of this model for human disease.
Here we assess the impact of four commonly used antibiotics on colonic microbiota and mucosal immunity in healthy RM as follows: (i) Cephalexin, a β-lactam antibiotic commonly used to treat a variety of Gram-positive bacterial infections including skin and soft tissue infections,23 and part of a class of antibiotics, first-generation cephalosporins, widely used as prophylactic antibiotics in surgical procecdures in macaques; (ii) Clindamycin, a lincosamide antibiotic used for a plethora of human conditions including acne, soft tissue infections, second-line strep throat treatment, bacterial vaginosis, and methicillin-resistant Staphylococcus aureus infection,24,25 and recognized by primate veterinarians for its potential to induce mild colitis; (iii) Paromomycin, an aminoglycoside antibiotic commonly used in HIV-infected individuals for Cryptosporidium infections26 and for its extremely low absorption, confining its effects to the GI tract; and (iv) Enrofloxacin is a fluoroquinolone antibiotic commonly used in veterinary practice and given often to NHPs prophylactically for medical studies.27 We demonstrate that all four antibiotics disrupted the native microbiota, leading to reduced concentrations of fecal SCFA, and that this was linked to an infiltration of neutrophils and interleukin (IL)-17-producing cells in the colonic mucosa. These data are the first to demonstrate the longitudinal effects of multiple antibiotic treatments on microbial composition, mucosal immunity, bacteria fermentation, inflammation, and MT.
Materials and methods
Study animals and antibiotic treatment
Animals were housed and cared for in Association for the Assessment and Accreditation of Laboratory Animal Care international-accredited facilities, and all animal procedures were performed according to protocols approved by the Institutional Animal Care and Use Committee of University of Washington (Protocol 4304-16). None of the animals included in this study received antibiotics within 6 months prior to the start of the study. Twelve female RMs were treated with antibiotics (n = 3/group) including the following: enrofloxacin (12 mg/kg, n = 3, once daily, 9 days), cephalexin (30 mg/kg, n = 3, once daily, 9 days), paromomycin (25 mg/kg, n = 3, twice daily, 9 days), or clindamycin (10 mg/kg, n = 3, twice daily, 6 days). We collected blood, biopsies of the mid-descending colon ~20–30 cm from the anus, and stool before, during, and after the antibiotic treatment according to the study schedule in (Fig. 1). Stool and two biopsies were stored at −80 °C immediately upon collection. We also stored one biopsy from each animal at each time point in RNALater solution. Blood and the remaining biopsies were processed immediately after collection as described below. None of the animals had any clinical complications related to the antibiotic treatment.
Animals (n = 3 per group) were treated with enrofloxacin, cephalexin, or paromomycin for 9 days or with clindamycin for 6 days. Two sets of samples were collected prior to the treatment. During the treatments, non-invasive samples were collected three times and mucosal samples collected once. Animals were tracked for 63 days after initiation of the antibiotic treatments.
DNA extraction, 16S rRNA gene sequencing, and data analysis
We extracted DNA from cryopreserved stool and colon biopsies using the PowerFecal DNA Isolation Kit (Qiagen, Valencia, CA). We then prepared sequencing libraries as described by the Earth Microbiome Project28 and sequenced them using the Illumina MiSeq Sequencer (Illumina, San Diego, CA). All sequence reads and operational taxonomic unit (OTU) observations were included in our analyses, to maximize the observed diversity of the bacterial communities. Sequencing data were analyzed using the QIIME software.29 We clustered OTUs at 97% similarity using the SWARM algorithm30 and assigned taxonomy based on sequence similarity to the SILVA database.31 We calculated alpha diversity using the Inverse Simpson Index, β-diversity using Bray–Curtis dissimilarity, and performed principal coordinates analysis (PCoA) using the ape and vegan packages in R. Sequences have been submitted to the NCBI SRA (Accession numbers for the datasets are GSE143729 and PRJNA604177).
Gas chromatography-mass spectrometry
We first weighted 0.05-0.1 g of stool into a sterile microcentrifuge tube and suspended the stool in acidified water (pH 2.5) at a concentration of 0.1 g/mL, vortexed thoroughly, and centrifuged at maximum speed for 10 min. We then transferred 250 µL of the supernatant to a fresh microcentrifuge tube and added 50 µL of an 800 µM solution of 2-methyl valeric acid in acidified water, which served as an internal standard. We next added 300 µL of ethyl acetate, vortexed for 2 min, and then centrifuged at maximum speed for 10 min. We then transferred 100 µL of the organic phase into an autosampler vial and analyzed SCFA concentrations on a Shimadzu QP2010 GCMS (Shimadzu, Kyoto, Japan). We used a Stabilwax fused silica column (30 m x 0.32 mm × 0.25 mm, Restek Bellefonte, PA) with helium as the mobile phase with a linear velocity of 47.2 cm/s. The method started at 90 °C and increased at a rate of 10 °C/min to 180 °C and then at a rate of 20 °C/min to 250 °C, and held at that temperature for 2 min. We used solutions of acetic, propionic, butyric, isobutyric, valeric, and isovaleric acids (Sigma Aldrich, St. Louis, MO) in ethyl acetate as calibration standards.
Blood and tissue processing
We collected blood in Vacutainer tubes (BD, Franklin Lakes, NJ) and transported immediately to the lab. We then centrifuged the whole blood at 2000 r.p.m. for 10 min at 24 °C to separate the cells and plasma, then collected 1 mL aliquots of plasma, and stored them at −80 °C. We stored biopsies in RPMI 1640 medium (GE Healthcare Life Sciences, Pittsburgh, PA) upon collection and immediately transported them to the laboratory. We stored two biopsies at −80 °C and one in RNALater solution as noted above for DNA extraction and RNA-sequencing (RNA-seq) analysis, respectively. We placed the remaining biopsies into 50 mL RPMI medium supplemented with 1× Pennicillin/Streptomycin (GE Healthcare Life Sciences), 40 µg/mL Liberase TL (Sigma Aldrich), and 4 µg/mL DNase (Sigma Aldrich), and vigorously stirred at 37 °C for 1 h. We then manually dissociated the digested biopsy tissue into a single-cell suspension by grinding them across a sterile 40 µm mesh filter. We then centrifuged the cells at 1900 r.p.m. for 6 min at 4 °C, suspended them in 8 mL RPMI 1640 supplemented with 10% fetal bovine serum, and 1× Pennicillin/Streptomycin (R10), and divided this suspension evenly between two 5 mL round-bottom tubes for flow cytometry analysis.
Antibody staining and flow cytometry
Innate and adaptive immunophenotyping
We stained one tube of the single-cell suspensions immediately after processing in each tissue and blood. We first washed the cells in 4 mL of sterile phosphate-buffered saline (PBS) and stained them with the LIVE/DEAD Fixable Aqua Dead Cell Stain Kit (ThermoFisher Scientific, Waltham, MA) for 5 min at room temperature. We then stained the cells with the following surface antigen antibodies, all of which were obtained from BD Biosciences (San Jose, CA), unless otherwise stated, for 20 min at 4 °C: CD45-PerCP (clone D058-1283), CD11b-APC-H7 (clone ICRF44), CCR6-Pe (clone 11A9), CD3-Pe-CF594 (clone SP34-2), CD20-Pe-Cy5 (clone 2H7), PD-1 (eBioscience, San Diego, CA, clone eBioJ105), CD274 (Biolegend, San Diego, CA, clone 29E.2A3), CD8-BV570 (Biolegend clone RPA-T8), CD4-BV605 (Biolegend clone OKT4), CD16-BV650 (clone 3G8), HLA-DR-BV711 (clone G46-6), and CD14-BV785 (clone M5E2). We next washed the cells in sterile PBS and permeabalized them using BD Cytofix/Cytoperm for 20 min at 4 °C. We then washed the cells two times in BD Perm/Wash buffer and stained them with the following intracellular antigen antibodies, all of which were obtained from BD Biosciences, unless otherwise stated, for 20 min at 4 °C: Ki67-FITC (clone B56), IL-23-eFLuor660 (eBioscience clone 23dcdp), Arginase-1-Ax700 (R&D Systems, Minneapolis, MN, clone 658922), and Caspase-3-V450 (clone C92-605). We then fixed the cells in 1% paraformaldehyde solution (Sigma Aldrich). Data are shown only for significantly altered parameters.
Intracellular cytokine staining
We stimulated the second tube of the single-cell suspension with 10 ng/mL phorbol myristate acetate (Sigma Aldrich) and 1 µg/mL ionomycin (Life Technologies) in 2 mL R10 supplemented with 1 mg/mL brefeldin A for 16 h at 37 °C. We then washed the cells with 4 mL sterile PBS and stained them with the LIVE/DEAD Fixable Aqua Dead Cell Stain Kit (ThermoFisher Scientific, Waltham, MA) for 5 min at room temperature. We then stained the cells with the following surface antigen antibodies, all of which were obtained from BD Biosciences (San Jose, CA), unless otherwise stated, for 20 min at 4 °C: CD45-PerCP (clone D058-1283), CD8-APC-H7 (clone SK1), CD3-PE-CF594 (clone SP34-2), CD11b-PE-Cy5 (clone ICRF44), CD11c-PE-Cy7 (eBioscience clone 3.9), CD20-BV570 (Biolegend clone 2H7), CD4-BV605 (Biolegend clone OKT4), HLA-DR-BV711 (clone G46-6), and CD14-BV785 (clone M5E2). We then washed the cells in sterile PBS and permeabalized them using BD Cytofix/Cytoperm for 20 min at 4 °C. We then washed the cells twice in BD Perm/Wash buffer and stained them with the following intracellular antigen antibodies, all of which were obtained from BD Biosciences, unless otherwise stated, for 20 min at 4 °C: IL6-FITC (MQ2-6A3), GM-CSF-APC (Biolegend clone BVD2-21C11), TNF-α-AF700 (clone Mab11), IL-17-Pe (eBioscience clone ebio64CAP17), IL-22-PerCP-eflour 710 (eBioscience clone IL022JOP), IL-8-BV421 (clone G265-8), and IFNγ-BV650 (Biolegend clone 4 S.B3). We then fixed the cells in 1% paraformaldehyde solution (Sigma Aldrich). We collected stained cells on an LSR II flow cytometer (BD Biosciences) and analyzed the data using FlowJo software (v10.2, FlowJo LLC, Ashland, OR).
RNA sequencing
We isolated total RNA from colonic biopsies stored in RNALater solution using Qiagen RNeasy Micro Kits (Qiagen, Valencia, CA). We then generated sequencing libraries using TruSeq Stranded Total RNA with Ribo-Zero Globin kits (Illumina, San Deigo, CA) and 200 ng of RNA as the input. We then sequenced the libraries on an Illumina HiSeq 2500 Sequencer (Illumina). We trimmed the raw sequence reads of adapters and filtered them using skewer to discard reads with an average phred quality score of <30 or a length of <36. Trimmed reads were then aligned using the HISAT232 aligner to the NCBI reference Mmul macaque genome (Mmul_8.0.1) and sorted using SAMtools.33 Aligned reads were counted and assigned to gene meta-features using featureCounts34 as part of the Subread package. Sequence and gene expression data are available at the Gene Expression Omnibus (Accession numbers for the datasets are GSE143729 and PRJNA604177).
Enzyme-linked immunosorbent assay
We used the Human soluble CD14 Quantikine ELISA Kit (R&D Systems, Minneapolis, MN) to quantify plasma concentrations of sCD14 before, during, and after the antibiotic treatment according to the manufacturer’s instructions. We diluted plasma 1:200 prior to analysis.
Statistical analyses
Given the similiarities in response to the different antibiotics classes, the 12 animals were grouped for statistical analysis longitudinally prior to and after antibiotic treatment. We conducted paired t-tests across all 12 animals to determine statistical differences between SCFA concentrations and immunological data, including flow cytometry and enzyme-linked immunosorbent assay. To evaluate statistical differences in bacterial abundances, we utilized the Wilcoxon’s signed-rank test using the abundance for each bacterial species at each time point. All statistics were corrected for multiple comparisons using a post-hoc Bonferroni correction. We performed these analyses using GraphPad Prism (v5.0 GraphPad, La Jolla, CA). For our RNA-seq analysis approach, we imported the count files into R and performed quality control, normalization, and analysis using the generalized linear model likelihood ratio test for pairwise differential gene expression testing between time points and treatments implemented in Bioconductor’s edgeR.35 Gene-set variation analysis36 was used for pathway-enrichment analysis with subsequent testing of significantly differentiated pathways using LIMMA’s37 moderated t-statistic. P-values for both the gene expression analysis and pathway enrichment were calculated with a maximum nominal P ≤ 0.05 chosen as significant. Longitudinal gene expression and pathway enrichment figures were generated using the ggplot2 library.38
Results
Antibiotics disrupt microbiota in the colonic mucosa and stool
We used 16S rRNA gene sequencing to profile the bacterial communities in the colonic mucosa and stool before, during, and after the antibiotic treatments. Prior to the antibiotic treatment (Days −42 and −21), the colon mucosa of all animals was dominated by Helicobacteraceae (average 67.1%, range 5.6%–99%), with minor populations of Ruminococcaceae (average 7%), Prevotellaceae (average 6.9%), Lachnospiraceae (average 2.7%), and Spirocheateceae (average 2.7%), in strong agreement with our and other groups’ data on the microbiota associated with the colon mucosa in RMs22,39 (Fig. 2a). Rapidly after antibiotic treatment (Day 3), there was a significant decrease in the abundance of the dominant Helicobacteriaceae (P = 0.0161) with increased abundances of Prevotellaceae and Ruminococcaceae, although there was no change in the abundance of these bacteria due to treatment with clindamycin. After the antibiotic treatments had ended (Days 14, 28, and 63), the mucosal bacterial communities generally returned to the pre-antibiotics state, with high abundances of Helicobacteriaceae, but the rate and extent of this recovery differed between the three groups that showed a response to the antibiotic treatment. Specifically, administration of paromomycin appeared to have longer-lasting effects than did the other antibiotics.
a Mucosal bacterial communities of all animals showed a high abundance of Helicobacteraceae prior to the antibiotic treatment. The antibiotics disrupted the mucosal bacterial communities, but clindamycin showed no impact on the predominant Helicobacteraceae. b Stool-associated bacterial communities of all of the animals were highly diverse, predominated by bacteria from the phyla Firmicutes and Bacteroides. Each antibiotic disrupted these communities, but the effects on community composition varied between the different antibiotics. c Random Forest analysis demonstrated that Enterobacteriaceae were the most strongly discriminating bacterial group in the stool bacterial communities at time points when antibiotics were being administered. d Other discriminating groups included Verrucomicrobiaceae, Helicobacteraceae, and Streptococcaeae, among others. The most abundant genera of Enterobacteriaceae included Escherichia, Proteus, and Enterobacter with a small proportion of Klebsiella. e The relative abundance of Enterobacteriaceae in the stool bacterial communities increased rapidly upon initiation of the antibiotic treatments (Day 0–7), persisted in some animals until Day 14, and then returned to pre-antibiotic levels by Day 49. f Akkermansia, detected in the stool bacterial communities, increased in relative abundance near the end of the antibiotic treatment and then persisted through the end of the study. g Ruminococcaceae displayed decreased relative abundance on Days 7 and 14, and increased back to pre-antibiotic levels on Day 28. e–g Green points indicate samples taken prior to the antibiotic treatment, red points indicate samples during the antibiotic treatment, and blue points indicate samples taken after cessation of the antibiotic treatment. Statistics were performed across all antibiotic treatment groups using a Wilcoxon’s signed-rank test and corrected for multiple comparisons using a post-hoc Bonferroni correction.
We next evaluated the stool-associated bacterial communities (Fig. 2b). Prior to the antibiotic treatment, all of the animals showed high abundances of Prevotellaceae and Ruminococcaceae among other, smaller populations of Firmicutes and Bacteroidetes bacteria. All four antibiotics disrupted composition of the stool microbiota, but these effects were less pronounced and more variable than those observed in the mucosa. Thus, we used the random forests classifier to determine which bacterial groups were most discriminative between time points with and without antibiotics. We found that the abundances of Enterobacteriaceae and Verrumicrobiaceae were the most discriminative between the three time points, followed by Succinivibrionaceae, Streptococcaceae, and Helicobacteriaceae (Fig. 2c). We evaluated the genera detected from the family Enterobacteriaceae and found that it was predominantly Escherichia, with lower abundances of Proteus, Enterobacter, and Klebsiella (Fig. 2d). The abundance of bacteria from the family Enterobacteriaceae was low before the antibiotic treatment, but rapidly increased during the antibiotic treatment, and remained increased following cessation of antibiotic treatments (Fig. 2e). We also found that the abundance of Verrucomicrobiaceae, which was determined to be an Akkermansia sp. was significantly increased during the antibiotic treatment and remained increased after the treatments ended (Fig. 2f). We also observed decreased abundances of Ruminococcaceae, one of the key types of fermentative bacteria, during the antibiotic treatment (Fig. 2g).
We evaluated species diversity in the mucosal- and stool-associated bacterial communities using the Inverse Simpson Index and found that there was no significant change in the diversity of the mucosal communities (Fig. 3a). However, there was a significant decrease in the diversity of the stool bacterial communities on Days 7 and 14, which returned to pre-antibiotic values by Day 28 (Fig. 3b). The perturbation to the colonic bacterial communities was further demonstrated by PCoA, in which samples taken during the antibiotic treatment tended to cluster separately from those taken before or after the antibiotic treatment (Fig. 3c, d), indicating disruption of the communities by the antibiotics. Together, these data demonstrate that the antibiotic treatments induced significant alterations to the diversity and composition of the bacterial communities in the colonic mucosa and lumen.
a Species diversity, as measured by the Inverse Simpson Index, was altered by antibiotic treatment across all animals in the four antibiotic treatment groups. The colonic mucosa showed increased species diversity during (Day 3) and shortly after (Day 14) the antibiotic treatments. b Stool communities showed reduced diversity during (Days 0, 3, 7) and shortly after (Day 14) the antibiotic treatment. c, d Principal coordinates analysis confirmed that the antibiotics disrupted the community composition in the mucosa c and stool d during the antibiotic treatment. Green points indicate samples taken prior to the antibiotic treatment, red points indicate samples during the antibiotic treatment, and blue points indicate samples taken after cessation of the antibiotic treatment. Statistics were performed across all groups using a Wilcoxon’s signed-rank test and corrected for multiple comparisons using a post-hoc Bonferroni correction.
Antibiotic treatment reduces stool SCFA concentrations
SCFA are end products of bacterial fermentation that play major roles in regulating host health and immunity.40 Thus, we used gas chromatography-mass spectrometry to measure the concentrations of acetate, propionate, butyrate, isobutyrate, valerate, and isovalerate in the stool of the RMs throughout the course of the study (Fig. 4). Before the antibiotic treatment, we detected all six SCFA in the range of 0.98–8.3 mM. Acetate was present in the highest concentrations (average 2.39 ± 1.11 mM), followed by butyrate (average 0.89 ± 0.7 mM), propionate (average 0.7 ± 0.86 mM), isovalerate (average 0.63 ± 0.37 mM), valerate (average 0.59 ± 0.47 mM), and isobutyrate (average 0.39 ± 0.21 mM). During the antibiotic treatment, the concentrations of all six SCFA significantly decreased as compared with baseline, but recovered to pre-antibiotic concentrations by Day 14. The observed reduction in SCFA concentrations in the stool was independent of the antibiotic used.
Across all four antibiotic treatment groups, stool concentrations of the SCFA acetate, propionate, butyrate, isobutyrate, valerate, and isovaerate were significantly reduced during the antibiotic treatment (indicated by gray bar) and rapidly returned to pre-antibiotic levels following cessation of the antibiotic treatments. Enrofloxacin—red circles, cephalexin—blue squares, paromomycin—green triangles, clindamycin—black upside-down triangles. The table below shows P-values that were determined using a paired t-test with Bonferroni’s correction for multiple comparisons (NS not significant). Statistics were performed across all antibiotic treatment groups.
Antibiotic treatment is associated with altered colonic mucosal immunity
We used multicolor flow cytometry to evaluate immune profiles of colon biopsies throughout the course of the study. We found that there were significantly increased frequencies of neutrophils (as previously defined by our group in RMs as CD45 + CD3-CD20-CD14 + CD11b + leukocytes41) during the antibiotic treatment that returned to normal levels after the antibiotic treatment had ended (Fig. 5a, b), and that the neutrophils expressed lower levels of Caspase-3 during the antibiotic treatment (Fig. 5c, d), indicating decreased levels of neutrophil apoptosis. This infiltration of neutrophils in the colonic mucosa was supported by RNA-seq data demonstrating an increase in CXCL8 expression (Fig. 5e), the coding gene for IL-8, as well as an overall increase in the expression of genes within the IL-8 pathway from Day 3–14, and began to return to pre-antibiotic levels by Day 28, showing strong temporal alignment with the flow cytometry data (Fig. 5f). However, not all of the treatment groups showed the same patterns of gene expression. Specifically, the enrofloxacin group showed a slight decrease in expression of CXCL8 and the IL-8 pathway on Day 3, whereas the other groups showed an increase in expression on that day.
a In all treatment groups, flow cytometry analysis demonstrated a significant increase of neutrophils in the colonic mucosa during the antibiotic treatment (indicated by the gray bar), which persisted to Day 14 and then returned to pre-antibiotic levels by Day 28–63. b Representative flow cytometry plots demonstrating accumulation of neutrophils on Day 3 of the antibiotic treatment. Neutrophils were specifically identified as CD45+CD3−CD20−CD14+CD11b+ leukocytes. CD11bHiCD14Hi cells were specifically gated out to exclude monocytes from the analysis. c, d We observed a significant decrease in Caspase-3+ neutrophils at days 14 and 28, which returned to pre-antibiotic levels by Day 63. e, f RNA-seq analysis demonstrated increased expression of cxcl8, the gene encoding IL-8 (e), as well as the IL-8 production pathway (f) with strong temporal alignment with the neutrophil infiltration observed through the flow cytometry analysis. Interestingly, the enrofloxacin group showed reduced expression of cxcl8 and the IL-8 production pathway on Day 3, but increased expression on Day 14. Enrofloxacin—red circles, cephalexin—blue squares, paromomycin—green triangles, clindamycin—black upside-down triangles. P-values were determined using a paired t-test with Bonferroni’s correction for multiple comparisons. Statistics were performed across all antibiotic treatment groups.
Specific bacteria and bacterial products in the GI tract can influence the polarization of CD4+ T-cell subsets.40 Thus, we hypothesized that the antibiotic-induced microbiome perturbations could affect frequencies of key CD4+ T-cell subsets. Although the frequency of CD4+ T cells in the colonic mucosa remained stable throughout the course of the study (data not shown), we observed increased frequencies of CD4+ T cells producing IL-17 (Fig. 6a, b) on Days 14, which reached significance by Day 28. Similarly, we observed increased frequencies of CD4+ T cells producing IL-22 (Fig. 6c) on Days 14 and 28. Using RNA-seq, we demonstrated that there was increased expression of genes in the IL-17 pathway for all treatment groups on Days 14–28, which returned to pre-antibiotic levels afterward (Fig. 6d).
a Flow cytometry analysis demonstrated an increase of IL-17-producing CD4+ T cells (defined as CD45+CD3+CD4+IL-17+) in the colonic mucosa after the antibiotic treatment (indicated by the gray bar) at Day 14 and a significant increase at Day 28, which returned to pre-antibiotic values by Day 63. b Representative flow cytometry plots demonstrating increased frequency of IL-17-producing CD4+ T cells on Day 28. c We observed an increase in the frequency of IL-22-producing CD4+ T cells. d mRNA-seq analysis similarly demonstrated increased expression of genes in the IL-17 pathway on Days 14–28 after the antibiotic treatment. Enrofloxacin—red circles, cephalexin—blue squares, paromomycin—green triangles, clindamycin—black upside-down triangles. P-values were determined using a paired t-test with Bonferroni’s correction for multiple comparisons. Statistics were performed across all antibiotic treatment groups.
To assess levels of systemic innate immune activation,42 we measured plasma concentrations of sCD14. We observed a minor, but significant increase in plasma sCD14 during the antibiotic treatment, which then returned to baseline values by Day 14 (Fig. 7). These data demonstrate that, during and after the antibiotic treatment, there were significant changes in mucosal immunity indicative of immune activation.
Plasma concentrations of sCD14 increased during antibiotic treatment. Across all treatment groups, we observed an increase in plasma concentrations of sCD14 during the antibiotic treatment (indicated by the gray bar), which reached significance at Day 7. Plasma sCD14 levels returned to pre-antibiotic values following cessation of antibiotics. These data support an inflammatory response during the antibiotic treatment. Enrofloxacin—red circles, cephalexin—blue squares, paromomycin—green triangles, clindamycin—black upside-down triangles. P-values were determined using a paired t-test with Bonferroni’s correction for multiple comparisons. Statistics were performed across all antibiotic treatment groups.
Discussion
Antibiotics are a cornerstone of modern medicine, but unrestricted use has led to a variety of problems, including increased incidence of antibiotic-resistant bacteria, as well as long-term alterations to the composition of colonic microbiota.43 Here we demonstrated that oral administration of antibiotics disrupted colon mucosal and stool-associated bacterial communities in healthy RMs and, importantly, demonstrate for the first time specific alterations in mucosal immunity after antibiotics.
Of the antibiotic-induced microbiome alterations, the most prominent was increased abundances of Enterobacteriaceae. Expansion of enteric pathogens after antibiotic treatment has been observed previously in murine models and is generally attributed to increased availability of carbohydrates after depletion of the native microbiota.44,45 A further explanation may also be related to antibiotic resistance. Indeed, antibiotic-resistant Enterobacteriaceae are a major emerging public health burden.46 We also observed increased abundances of Akkermansia during the antibiotic treatment. Akkermansia muciniphila is a well-characterized intestinal mucolytic bacterium47 and these bacteria have previously been shown to increase in abundance following antibiotic treatments.18 Whether or not the increased abundance of Akkermansia was due to altered substrate pools is unclear; however it is important to note that increased abundances of mucolytic bacteria may alter mucous properties and thus may affect the integrity of the mucosal layers, and further studies understanding mechanisms of altered microbes after antibiotics are warranted.
Although the effects on the microbiota varied between the different antibiotics, all four antibiotics elicited a dramatic reduction in stool SCFA concentrations. There are several explanations for this phenomenon. One explanation is that the antibiotics altered the bacterial metabolism, stalling SCFA production. Antibiotics are known to induce changes in bacterial metabolism and, although some of these responses are specific to the type of antibiotic used, others appear to be more generalized across multiple classes of antibiotics.48,49 Such metabolic alterations induced by antibiotics, combined with continued utilization of SCFAs by the microbiota and host, may help explain the observed loss of SCFA production in this study. Recent evidence has demonstrated that propionate produced by commensal Bacteroides spp. inhibits the growth of pathogenic Salmonella typhimurium in murine models.50 Thus, the decreased stool SCFA concentrations in this study may have played a role in the expansion of Enterobacteraceae spp. However, further investigation is necessary to determine the precise cause and impacts of the decreased SCFA.
The immunological responses to the antibiotic-induced microbiome disruption also appear to be conserved across the different antibiotic classes. Indeed, we observed increased frequencies of neutrophils, Th17, and Th22 cells by flow cytometry, either during or after the antibiotic treatment and these responses were conserved across all of the study groups. These alterations in mucosal immune status were further supported by RNA-seq, demonstrating increased expression of genes in the IL-8 and IL-17 pathways. Neutrophils are recruited to the sites of bacterial infections and are critical for protecting against pathogenic bacteria.51 In the context of this study, it is possible that neutrophils were recruited to the colonic mucosa in response to the altered microbiota composition. We also demonstrated reduced neutrophil apoptosis during the antibiotic treatment. Increased neutrophil lifespan may contribute to mucosal inflammation through increased release of antimicrobial factors such as proteases and reactive oxygen species, which can disrupt the integrity of mucosal epithelial barriers.52 This is supported by the increased plasma concentrations of sCD14 post treatment, which has been extensively used as a biomarker of reduced mucosal epithelial integrity and translocation of microbes and microbial products from the intestinal lumen into the periphery.3
Neutrophils have also been shown to play a role in recruiting Th17 cells53 and this is supported by the timing of the increase of Th17 cells and global gene expression patterns in our study. Th17 cells are important for maintaining mucosal homeostasis and for protection against intestinal pathogens.54 Thus, the observed increase in Th17 cells in this study was likely also in response to the antibiotic-induced microbiota alterations. However, it is also important to note that approximately at the same time that we observed increased Th17 cells (D14-D28), we also observed increased frequencies of Th22 cells, as well as CD4+ T cells producing both IL-17 and IL-22. Th22 cells (and IL-22) are important for protecting against pathogenic bacteria,55 but also promote epithelial regeneration and repair.56 Thus, the observed increase of Th17 and Th22 cells could also be in response to mucosal dysfunction and thus important for restoration of the colonic epithelium during the resolution of inflammation caused by the influx of neutrophils. The precise mechanisms underlying these changes in mucosal immunity during antibiotic treatment require further study.
A caveat of these studies is the very small group size in each antibiotic category. This study was initially designed as a pilot study to determine which antibiotics should be focused on for NHP research. Given the difference in mechanisms of activity of each antibiotic, it was somewhat surprising how conserved the microbial and immune responses to all the groups was. Thus, we chose to group all antibiotics for analysis to focus on these conserved mechanisms. Subsequent studies focused on individual antibiotics and determining whether potential disparate effects would be seen in larger treatment groups will be important for future studies.
In summary, these data demonstrate that antibiotic treatments disrupt colonic bacterial communities and promote mucosal inflammation and epithelial disruption in healthy RMs. Macaques are the most biologically relevant animal model for studies of human immunology and are used as a model for a variety of diseases, in particular HIV infection.9,57 HIV and pathogenic SIV infection are associated with dysbiosis of the intestinal microbiota and significant dysfunction of the mucosa independent of antibiotics.12,58,59 Thus, antibiotic use in HIV-infected individuals might exacerbate these conditions and contribute to further immune activation. Further, antibiotic use in studies of SIV infection that involve evaluations of mucosal immunity and the intestinal microbiota can have dramatic impacts on the results. These data suggest that antibiotic use should be avoided unless absolutely necessary in such studies.
Change history
24 February 2020
A Correction to this paper has been published: https://doi.org/10.1038/s41385-020-0274-x
References
Thaiss, C. A., Levy, M., Suez, J. & Elinav, E. The interplay between the innate immune system and the microbiota. Curr. Opin. Immunol. 26, 41–48 (2014).
Hullar, M. A. & Fu, B. C. Diet, the gut microbiome, and epigenetics. Cancer J. 20, 170–175 (2014).
Brenchley, J. M. et al. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat. Med. 12, 1365–1371 (2006).
Dillon, S. M. et al. Gut dendritic cell activation links an altered colonic microbiome to mucosal and systemic T-cell activation in untreated HIV-1 infection. Mucosal Immunol. 9, 24–37 (2016).
Jiang, W. et al. Plasma levels of bacterial DNA correlate with immune activation and the magnitude of immune restoration in persons with antiretroviral-treated HIV infection. J. Infect. Dis. 199, 1177–1185 (2009).
Sandler, N. G. et al. Plasma levels of soluble CD14 independently predict mortality in HIV infection. J. Infect. Dis. 203, 780–790 (2011).
Vujkovic-Cvijin, I. et al. Dysbiosis of the gut microbiota is associated with HIV disease progression and tryptophan catabolism. Sci. Transl. Med. 5, 193ra191 (2013).
Gardner, M. B. & Luciw, P. A. Macaque models of human infectious disease. ILAR J. 49, 220–255 (2008).
Messaoudi, I., Estep, R., Robinson, B. & Wong, S. W. Nonhuman primate models of human immunology. Antioxid. Redox Signal. 14, 261–273 (2011).
Veazey, R. S. & Lackner, A. A. Nonhuman primate models and understanding the pathogenesis of HIV infection and AIDS. ILAR J. 58, 160–171 (2017).
Ericsen, A. J. et al. Microbial translocation and inflammation occur in hyperacute immunodeficiency virus infection and compromise host control of virus replication. PLoS Pathog. 12, e1006048 (2016).
Handley, S. A. et al. SIV infection-mediated changes in gastrointestinal bacterial microbiome and virome are associated with immunodeficiency and prevented by vaccination. Cell Host Microbe. 19, 323–335 (2016).
Handley, S. A. et al. Pathogenic simian immunodeficiency virus infection is associated with expansion of the enteric virome. Cell 151, 253–266 (2012).
Klase, Z. et al. Dysbiotic bacteria translocate in progressive SIV infection. Mucosal Immunol. 8, 1009–1020 (2015).
Moeller, A. H. et al. SIV-induced instability of the chimpanzee gut microbiome. Cell Host Microbe. 14, 340–345 (2013).
Evans, T. I. et al. SIV-induced translocation of bacterial products in the liver mobilizes myeloid dendritic and natural killer cells associated with liver damage. J. Infect. Dis. 213, 361–369 (2016).
Hensley-McBain, T. et al. Effects of fecal microbial transplantation on microbiome and immunity in Simian immunodeficiency virus-infected macaques. J. Virol. 90, 4981–4989 (2016).
Dubourg, G. et al. High-level colonisation of the human gut by Verrucomicrobia following broad-spectrum antibiotic treatment. Int. J. Antimicrob. Agents 41, 149–155 (2013).
Knoop, K. A. et al. Antibiotics promote the sampling of luminal antigens and bacteria via colonic goblet cell associated antigen passages. Gut Microbes 8, 400–411 (2017).
Knoop, K. A., McDonald, K. G., Kulkarni, D. H. & Newberry, R. D. Antibiotics promote inflammation through the translocation of native commensal colonic bacteria. Gut. 65, 1100–1109 (2016).
Candon, S. et al. Antibiotics in early life alter the gut microbiome and increase disease incidence in a spontaneous mouse model of autoimmune insulin-dependent diabetes. PLoS ONE 10, e0125448 (2015).
Zevin A. S., et al. Antibiotic treatment disrupts bacterial communities in the colon and rectum of simian immunodeficiency virus-infected macaques. FEMS Microbiol Lett 364, fnx228 (2017). https://doi.org/10.1093/femsle/fnx228.
Kalman, D. & Barriere, S. L. Review of the pharmacology, pharmacokinetics, and clinical use of cephalosporins. Tex. Heart Inst. J. 17, 203–215 (1990).
Dhawan, V. K. & Thadepalli, H. Clindamycin: a review of fifteen years of experience. Rev. Infect. Dis. 4, 1133–1153 (1982).
Lell, B. & Kremsner, P. G. Clindamycin as an antimalarial drug: review of clinical trials. Antimicrob. Agents Chemother. 46, 2315–2320 (2002).
Caccio, S., Pinter, E., Fantini, R., Mezzaroma, I. & Pozio, E. Human infection with Cryptosporidium felis: case report and literature review. Emerg. Infect. Dis. 8, 85–86 (2002).
Kim, J. et al. Antimicrobial use for and resistance of zoonotic bacteria recovered from nonhuman primates. Comp. Med. 67, 79–86 (2017).
Walters W., et al. Improved bacterial 16S rRNA gene (V4 and V4-5) and fungal internal transcribed spacer marker gene primers for microbial community surveys. mSystems 1, pii: e00009-15 (2016).
Caporaso, J. G. et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336 (2010).
Mahe, F., Rognes, T., Quince, C., de Vargas, C. & Dunthorn, M. Swarm: robust and fast clustering method for amplicon-based studies. PeerJ. 2, e593 (2014).
Pruesse, E. et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35, 7188–7196 (2007).
Kim, D., Langmead, B. & Salzberg, S. L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Hanzelmann, S., Castelo, R. & Guinney, J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 14, 7 (2013).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Wickham, H. Ggplot2: Elegant Graphics for Data Analysis viii, 212 (Springer, New York, 2009).
Yasuda, K. et al. Biogeography of the intestinal mucosal and lumenal microbiome in the rhesus macaque. Cell Host Microbe. 17, 385–391 (2015).
Park, J. et al. Short-chain fatty acids induce both effector and regulatory T cells by suppression of histone deacetylases and regulation of the mTOR-S6K pathway. Mucosal Immunol. 8, 80–93 (2015).
Hensley-McBain, T. et al. Intestinal damage precedes mucosal immune dysfunction in SIV infection. Mucosal Immunol. 11, 1429–1440 (2018).
Shive, C. L., Jiang, W., Anthony, D. D. & Lederman, M. M. Soluble CD14 is a nonspecific marker of monocyte activation. Aids 29, 1263–1265 (2015).
Korpela, K. et al. Intestinal microbiome is related to lifetime antibiotic use in Finnish pre-school children. Nat. Commun. 7, 10410 (2016).
Faber, F. et al. Host-mediated sugar oxidation promotes post-antibiotic pathogen expansion. Nature 534, 697–699 (2016).
Ng, K. M. et al. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature 502, 96–99 (2013).
Savard, P. & Perl, T. M. A call for action: managing the emergence of multidrug-resistant Enterobacteriaceae in the acute care settings. Curr. Opin. Infect. Dis. 25, 371–377 (2012).
van Passel, M. W. et al. The genome of Akkermansia muciniphila, a dedicated intestinal mucin degrader, and its use in exploring intestinal metagenomes. PLoS ONE 6, e16876 (2011).
Belenky, P. et al. Bactericidal antibiotics induce toxic metabolic perturbations that lead to cellular damage. Cell Rep. 13, 968–980 (2015).
Zampieri, M., Zimmermann, M., Claassen, M. & Sauer, U. Nontargeted metabolomics reveals the multilevel response to antibiotic perturbations. Cell Rep. 19, 1214–1228 (2017).
Jacobson, A. et al. A gut commensal-produced metabolite mediates colonization resistance to Salmonella infection. Cell Host Microbe. 24, 296–307.e297 (2018).
Wera O., Lancellotti P., Oury C. The dual role of neutrophils in inflammatory bowel diseases. J. Clin. Med. 5, pii: E118 (2016).
Elbim, C. et al. Early divergence in neutrophil apoptosis between pathogenic and nonpathogenic simian immunodeficiency virus infections of nonhuman primates. J. Immunol. 181, 8613–8623 (2008).
Pelletier, M. et al. Evidence for a cross-talk between human neutrophils and Th17 cells. Blood 115, 335–343 (2010).
Wang, Z. et al. Regulatory T cells promote a protective Th17-associated immune response to intestinal bacterial infection with C. rodentium. Mucosal Immunol. 7, 1290–1301 (2014).
Basu, R. et al. Th22 cells are an important source of IL-22 for host protection against enteropathogenic bacteria. Immunity 37, 1061–1075 (2012).
Kim, C. J. et al. A role for mucosal IL-22 production and Th22 cells in HIV-associated mucosal immunopathogenesis. Mucosal Immunol. 5, 670–680 (2012).
Hatziioannou, T. & Evans, D. T. Animal models for HIV/AIDS research. Nat. Rev. Microbiol. 10, 852–867 (2012).
Brenchley, J. M. Mucosal immunity in human and simian immunodeficiency lentivirus infections. Mucosal Immunol. 6, 657–665 (2013).
Dillon, S. M. et al. An altered intestinal mucosal microbiome in HIV-1 infection is associated with mucosal and systemic immune activation and endotoxemia. Mucosal Immunol. 7, 983–994 (2014).
Acknowledgements
We thank all veterinary staff of the Washington National Primate Research Center for conducting the animal studies. These studies, in part, were funded with support from the WaNPRC NIH core grant P51 OD010425-51 and partially by NIH grants 1R01AI117828 and 1R01DK112254 to N.R.K. and amfAR 109222-58-RGRL to C.M.C. J.A.M. was supported by K01OD024876. We also thank the Case Western Applied Functional Genomics Core.
Author information
Authors and Affiliations
Contributions
Study design: N.R.K., J.S., and A.S.Z. Animal studies: J.S., D.M., S.W., and B.A. Sample processing: A.S.Z., J.A.M., C.M., E.C., T.G., and A.T.G. Experimental analysis: A.S.Z., J.A.M., R.C., T.H.M., C.M., A.T.G., B.R., M.F., M.C., and E.S. Data analysis: J.A.M., A.S.Z., T.H.M., M.C., C.M.C., M.J.C., and M.G. Consulting on clinical antibiotics use: H.B.H. Manuscript preparation and editing: J.A.M., A.S.Z., H.B.H., M.J.C., and N.R.K.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Manuzak, J.A., Zevin, A.S., Cheu, R. et al. Antibiotic-induced microbiome perturbations are associated with significant alterations to colonic mucosal immunity in rhesus macaques. Mucosal Immunol 13, 471–480 (2020). https://doi.org/10.1038/s41385-019-0238-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41385-019-0238-1