Abstract
Tumors of purported specialized prostatic stromal origin comprise prostatic stromal sarcomas (PSS) and stromal tumors of uncertain malignant potential (STUMP). Prior studies have described their clinicopathologic characteristics, but the molecular features remain incompletely understood. Moreover, these neoplasms are morphologically heterogeneous and the lack of specific adjunctive markers of prostatic stromal lineage make precise definition more difficult, leading some to question whether they represent a specific tumor type. In this study, we used next-generation DNA and RNA sequencing to profile 25 primary prostatic mesenchymal neoplasms of possible specialized prostatic stromal origin, including cases originally diagnosed as PSS (11) and STUMP (14). Morphologically, the series comprised 20 cases with solid architecture (11 PSS and 9 STUMP) and 5 cases with phyllodes-like growth pattern (all STUMP). Combined DNA and RNA sequencing results demonstrated that 19/22 (86%) cases that underwent successful sequencing (either DNA or RNA) harbored pathogenic somatic variants. Except for TP53 alterations (6 cases), ATRX mutations (2 cases), and a few copy number variants (−13q, −14q, −16q and +8/8p), the findings were largely nonrecurrent. Eight gene rearrangements were found, and 4 (NAB2-STAT6, JAZF1-SUZ12, TPM3-NTRK1 and BCOR-MAML3) were useful for reclassification of the cases as specific entities. The present study shows that mesenchymal neoplasms of the prostate are morphologically and molecularly heterogeneous and include neoplasms that harbor genetic aberrations seen in specific mesenchymal tumors arising in other anatomic sites, including soft tissue and the uterus. These data suggest that tumors of purported specialized prostatic stromal origin may perhaps not represent a single diagnostic entity or specific disease group and that alternative diagnoses should be carefully considered.
Similar content being viewed by others
Introduction
Tumors of purported specialized prostatic stromal origin are rare and comprise a wide spectrum of lesions with variable morphology and clinical behavior, including tumors of uncertain biologic potential [so-called “stromal tumors of uncertain malignant potential” (STUMP)] as well as overtly malignant neoplasms [“prostatic stromal sarcomas” (PSS)] [1]. PSS and STUMP are diagnostic categories that group mesenchymal neoplasms of the prostate that are not classifiable as more specific tumor types. However, it is uncertain whether they represent a distinct biological entity as their underlying molecular alterations remain incompletely understood. The lack of specific markers and the morphologic variability of tumors of purported specialized prostatic stromal origin suggests that they might be biologically and molecularly heterogeneous.
In the largest series, STUMP were three times more frequent than PSS [1]. The mean age at diagnosis was 58 years and patients usually presented with symptoms secondary to local mass effect and hematuria [1]. STUMP usually have an indolent clinical course without progression even after nonradical resection or during active surveillance [1, 2]. However, a significant proportion of STUMP has aggressive behavior characterized by local recurrences and the presence of synchronous or metachronous PSS. Four different growth patterns of STUMP were recognized initially [2]: (1) hypercellular stroma with scattered atypical cells intermingled with benign glands (2) hypercellular stroma without atypia intermingled with benign glands (3) hypercellular stroma, with or without atypia, associated with benign glands in a “leaf-like” configuration (phyllodes-like), and (4) hypercellular stroma without atypia that is not intermingled with benign glands. A later series identified additional cases with extensively myxoid stroma and without significant cytologic atypia [1].
The distinction between STUMP and PSS is not always possible based on morphology. Moreover, ~50% of PSS are associated with STUMP [1] and one of these components may not be represented adequately in core needle biopsies or transurethral resections. In a study published by Herawi and Epstein, the authors described that 7 of 14 PSS were associated with STUMP, either as metachronous or synchronous findings [1]. The metachronous association is characterized by an initial diagnosis of STUMP, followed by recurrence as PSS after treatment or progression to PSS during follow-up [1]. Alternatively, STUMP and PSS components can be synchronously present in the same specimen [1]. The latter scenario likely represents the progression/dedifferentiation of a neoplasm of low malignant potential that would otherwise have an indolent clinical course, as seen in other soft tissue tumors [3,4,5,6].
Although there are no clinically validated diagnostic criteria, earlier series have described a set of histologic features that can be used to distinguish STUMP from PSS (Table 1). In general, prominent mitotic activity, tumor necrosis, and overt stromal overgrowth are worrisome histologic findings, and the identification of two or more of these features is highly suggestive of PSS [2]. The microscopic appearance of PSS is variable and includes tumors with spindled and epithelioid cells arranged in fibrosarcomatous, storiform, and phyllodes-like growth patterns [1, 7], among others.
The molecular characteristics of prostatic stromal neoplasms remain somewhat unclear. Thus far, only 2 overlapping series of 19 and 14 cases (including both STUMP and PSS) have been studied with array comparative genomic hybridization, fluorescence in situ hybridization [8], and whole-exome DNA sequencing [9]. Both studies found frequent copy number variants (CNVs), with losses of chromosomes 13, 14, and 10 being the only recurrent findings [8, 9]. The overall tumor mutational burden was higher in PSS than in STUMP, but cancer-relevant mutations and structural variants were not detected [9].
At the present time, it is uncertain if tumors of purported specialized prostatic stromal origin are biologically and genetically homogeneous. As mentioned above, their widely variable morphology and the absence of specific lineage markers suggest that they are probably heterogeneous and might not represent a distinct entity. To further explore this premise, we analyzed a multi-institutional series of mesenchymal tumors of the prostate diagnosed as PSS and STUMP using DNA and RNA sequencing.
Materials and methods
This research was performed with the approval of the Institutional Review Board of Partners Healthcare (BWH).
Identification, accrual and review of cases
The pathology databases of four institutions (BWH, Vanderbilt University Medical Center, Cleveland Clinic, Advocate Lutheran General Hospital) and the personal consultation files of the authors (CDF and JKM) were queried for primary prostatic mesenchymal tumors of probable prostatic stromal origin, originally diagnosed as STUMP or PSS. Archival stained slides [H&E and immunohistochemistry (IHC)] and unstained slides were retrieved for the study. When available, additional 5-µm sections were cut from archival formalin-fixed paraffin-embedded tissue blocks. The original pathology reports and slides were first reviewed by the authors at the participating institutions (CDF, AMA, JKM, JBG, and MRP), and all cases included in the study were subsequently centrally reviewed at BWH (CDF and AMA).
DNA sequencing
A 447-gene NGS panel (OncoPanel) was performed as previously described by our group [10, 11]. Briefly, lesional areas with adequate cellularity (>20% tumor nuclei) were dissected manually using an H&E slide marked by a pathologist as a guide. DNA extraction was performed using a standard commercial kit (Qiagen, Valencia, CA) according to the manufacturer’s recommendations. After DNA shearing by sonication, 200 ng of DNA per sample (threshold of 100 ng) were used to prepare the sequencing libraries (TruSeq LT library preparation kit; Illumina, San Diego, CA). Sequences of interest were selected by hybridization to a set of custom-designed probes (Agilent SureSelect; Agilent Technologies, Santa Clara, CA) and sequencing was performed on the Illumina HiSeq 2500 System (Illumina, San Diego, CA). A validated institutional informatic pipeline was used for deconvolution of samples, read alignment, and calling/annotation of single nucleotide variant (SNVs), indels, CNVs and structural variants [10,11,12]. Also, mutational signatures (POLE, APOBEC, smoking, UV) and mismatch repair status were evaluated with validated in-house developed algorithms [13]. OncoPanel is a tumor-only assay; therefore, variants with a populational frequency >0.1% in the gnomAD database (Broad Institute) were filtered out to reduce contamination of sequencing results with germline variants. All reported variants were further reviewed and tiered for actionability/biologic relevance by a molecular pathologist (LMS). The variants were interpreted as possible drivers if they were biologically relevant and had a known driver role in other tumor types.
RNA sequencing
RNA sequencing for detection of gene fusion products was performed as previously described by Dickson et al. [14]. Briefly, tumor tissue was manually dissected from 2 to 5 FFPE tissue sections per case. Extraction and preparation of RNA were performed with a commercial kit following the manufacturer’s recommendation (ExpressArt FFPE Clear RNA Ready kit, Amsbio, Cambridge, MA). Subsequently, total RNA was assessed (Qubit RNA HS Assay kit, Thermofisher Scientific, Mississauga, ON) and sequencing libraries were prepared (TruSight RNA Fusion Panel, Illumina, San Diego, CA) using a net input of 20–100 ng RNA per case. Samples were sequenced with 76 base-pair paired-end reads on a MiSeq platform (Illumina, San Diego, CA) at 8 samples per flow cell producing a total of approximately 3 million reads per sample. Two informatic pipelines were used to analyze the results: STAR aligner with Manta fusion caller through the Illumina Local Run Manager (v.1.3.0), and BOWTIE2 alignment with the JAFFA fusion caller [15, 16]. Fusions were considered likely stochastic if they had previously been annotated in our molecular database as occurring in the context of another established driver, failed to maintain the reading frame, were non-exonic, or were low-confidence calls with few supporting reads [14].
Fluorescence in situ hybridization (FISH)
Two cases with CIC rearrangements detected by RNA sequencing were further evaluated by FISH (see the “Results” section below). Formalin-fixed paraffin-embedded tissue sections (4 µm) were evaluated using in-house developed, clinically validated dual-color break-apart CIC probes specific for the 5′ and 3′ regions of CIC at 19q13.2. Labeling and hybridization of the probes were performed according to standard protocols in our laboratory. FISH results were evaluated in tumor areas previously marked by a pathologist. Fifty nonoverlapping interphase tumor nuclei with distinct nuclear boundaries were assessed and scored manually by a cytogeneticist (PDC). Split signals were defined as those separated by one or more probe lengths. Cases were considered positive for CIC rearrangements when more than 2% (>1/50) of tumor nuclei harbored split signals.
Results
General characteristics of the cases
The series comprised 25 primary mesenchymal tumors of the prostate (20 resections and 5 biopsies) collected between 2000 and 2021 from 25 individual patients, with a median age at diagnosis of 60 years (range 17–78 years). The original diagnoses were PSS in 11 cases and STUMP in 14 cases (Table 2). For one of the STUMP (case 15), an intraperitoneal metastasis with identical morphology was subsequently received in consultation at BWH. Also, one of the cases counted as a PSS above (case 11) was associated with adjacent STUMP components, as previously described in the literature [1]. Although the two histologic components of the sample were differentially dissected for DNA sequencing, the case was considered a PSS for the purposes of the histopathologic evaluation presented below.
Microscopically, 20 cases (20/25, 80%; 11 PSS and 9 STUMP) had solid architecture and 5 cases (5/25, 20%; all STUMP) had at least focal phyllodes-like growth pattern. Two of the tumors with solid histology had a striking microscopic resemblance to low-grade endometrial stromal sarcoma characterized by hypercellular sheets of monotonous small round to oval cells with minimal atypia, bland round or ovoid nuclei, scant cytoplasm, and easily identifiable mitotic figures. Plexiform vascularity with a rich network of small arterioles and prominent capillaries was noted in both cases, and a partially tubular architecture (reminiscent of admixed sex-cord elements) was observed in one case.
Infiltration between benign prostatic glands was seen in 10 cases (10/25, 40%; 4 PSS and 6 STUMP), including all tumors with phyllodes-like morphology. Mitotic activity was highly variable, with a median number of mitoses of 1 per 10 hpf overall (range <1–25 per 10 hpf), 22 per 10 hpf for PSS (range 6–25 per 10 hpf), and <1 per 10 hpf for STUMP (range < 1–2 per 10 hpf). Twelve cases (12/25, 48%; all STUMP), including the tumor that metastasized mentioned above (case 15), had <1 mitosis per 10 hpf. All tumors were hypercellular compared to the normal prostatic stroma, with mild hypercellularity in 9 cases (9/25, 36%; 1 PSS, 8 STUMP), moderate hypercellularity in 7 cases (7/25, 28%; 2 PSS and 5 STUMP) and marked hypercellularity in 9 cases (9/25, 36%; 8 PSS and 1 STUMP). Necrosis was present in 6 cases (6/25, 24%; 4 PSS and 2 STUMP) and absent in the remaining 19 (19/25, 76%; 7 PSS and 12 STUMP). Nuclear atypia was absent in most cases (15/25, 60%; 7 PSS and 8 STUMP), mild in 4 cases (4/25, 16%; 2 PSS and 2 STUMP) and moderate in 6 cases (6/25, 24%; 3 PSS and 3 STUMP). The nuclear atypia seen in these tumors was mostly degenerative in type, characterized by homogeneously hyperchromatic smudgy chromatin. Multinucleate tumor cells were identified in 7 cases (7/25, 28%; 2 PSS and 5 STUMP) and absent in the remaining 18 cases (18/25, 72%; 9 PSS and 9 STUMP).
The histologic patterns of each tumor were also classified using previous definitions from the literature to enable comparison to molecular findings (Tables 2 and 3) [1]. For the 14 cases originally diagnosed as STUMP, the patterns present were as follows: hypercellular (5), phyllodes (5), degenerative atypia (1), “patternless” (1), and myxoid (2). The patterns in sarcomas were often admixed (i.e., not mutually exclusive) and included: short fascicles (5), patternless (5), epithelioid (4), storiform (3), and fibrosarcomatous (1).
IHC for keratins was performed in 7 cases (Pan-keratin in 4 cases, AE1/AE3 in 1 case, and AE1/AE3 plus CAM5.2 in 2 cases). Of these, only 1 case (1/7, 14%) had scattered cells positive for AE1/AE3, while the remaining 6 cases (6/7, 86%) were negative for keratins. EMA was focally or multifocally expressed in 2 of 6 cases (33%), but diffuse positivity was not seen in any case. Desmin, SMA, and CD34 were positive in 16/19 cases (84%), 10/16 cases (63%), and 5/10 cases (50%), respectively. STAT6 was considered negative for diagnostic purposes (i.e., not supportive of solitary fibrous tumor) in 5/5 cases (100%). However, an old case from 2003 with striking microscopic resemblance to low-grade endometrial stromal sarcoma (case 1, positive for NAB2-STAT6 fusion, see below) had weak multifocal nuclear STAT6 expression. Finally, PR positivity was seen in 12/19 cases (63%).
Illustrative micrographs of representative cases annotated with the corresponding genetic variants identified (see DNA sequencing and RNA sequencing results below) are presented in Figs. 1–5.
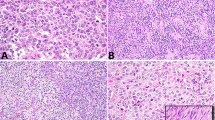
A Micrograph of case 1 (PSS). Despite its morphologic appearance, which was reminiscent of low-grade endometrial stromal sarcoma, this case harbored a NAB2-STAT6 fusion. B Case 12 (PSS). This case also had morphologic features suggestive of low-grade endometrial stromal sarcoma with partly tubular (sex-cord stromal tumor-like) architecture and harbored a JAZF1-SUZ12 rearrangement. Abbreviations: PSS prostatic stromal sarcoma.
A Case 2 (PSS) had minimal atypia, but both cellularity and mitotic activity were prominent. This case had a TPM3-NTRK1 rearrangement. B Case 3 (PSS) had a primitive appearance with small spindled and epithelioid cells and harbored a BCOR-MAML3 gene fusion. C Case 13 (STUMP) had a phyllodes-like architecture with myxoid stromal change and mild atypia. This case had a novel RGPD2-FOXO1 rearrangement. Abbreviations: STUMP stromal tumor of uncertain malignant potential, PSS prostatic stromal sarcoma.
A Case 4 (PSS) had marked nuclear atypia with multinucleate tumor cells. This case had biallelic inactivation of TP53, an ATRX mutation, and multiple copy number changes. B Case 16 (STUMP) had molecular evidence of biallelic TP53 inactivation. Microscopically, it was a spindle cell lesion with moderate nuclear atypia, inconspicuous mitotic activity, and scattered multinucleate tumor cells. C Case 14 had a TP53 mutation and a TP53 rearrangement involving the first intron of the gene. This was also a spindle cell lesion with mild hypercellularity, moderate nuclear atypia, and scattered multinucleate tumor cells. As seen in these cases, nuclear pleomorphism appears to be common in mesenchymal tumors of the prostate with TP53 aberrations.
A The copy number plot demonstrates focal high-grade amplification of the region of chromosome 12q that encompasses MDM2, CDK4, and ERBB3, without additional copy number changes. B, C The simple prostatectomy (enucleation) specimen showed a hypercellular neoplasm with spindle and epithelioid cells, no atypia, and low mitotic activity. Abbreviations: STUMP stromal tumor of uncertain malignant potential, PSS prostatic stromal sarcoma.
A At low magnification, three different areas of the tumor are identified: (1) an area of low cellularity and myxoid stroma (upper left), (2) an area with intermediate cellularity and denser stroma with intermingled benign prostate glands (middle), and (3) an area with high cellularity and a minimal amount of stroma (lower right). B The upper left area shown in A corresponds to a histologically bland component of the neoplasm which lacks mitoses and atypia and has a prominent myxoid stroma (“myxoid STUMP”). The STUMP component of the tumor harbored a CHEK2 mutation (possibly germline) and evidence of subclonal biallelic TP53 inactivation. C Areas of transition with increased cellularity and noticeable nuclear atypia (such as the one shown in the middle portion of micrograph A) were identified. D The micrograph illustrates a representative area of the high-grade sarcoma present in this case (PSS, lower right portion of micrograph A), which was characterized by epithelioid and spindled cells arranged in a patternless fashion. Marked nuclear atypia and a prominent mitotic activity were noted. The PSS component of the tumor harbored biallelic inactivation of both TP53 and CHEK2, as well as widespread LOH in the context of multiple copy number changes (see Fig. 7). Abbreviations: LOH loss of heterozygosity, STUMP stromal tumor of uncertain malignant potential, PSS prostatic stromal sarcoma.
DNA sequencing
Twenty-one samples (21/25, 84%) underwent successful sequencing, while the remaining 4 (4/25, 16%) either failed sequencing or did not yield sufficient DNA. The median tumor mutational burden was 5.7 mutations per Mb overall (range 1.5–9.9 mutations per Mb), 6.1 mutations per Mb for PSS (range 4.6–9.9 mutations per Mb) and 3.0 mutations per Mb for STUMP (range 1.5 to 9.1 mutations per Mb).
Eight gene rearrangements were found (Fig. 6), including 5/8 gene rearrangements with both partners identified and 3/8 gene rearrangements with intergenic regions or only 1 partner identified; however, recurrent structural events were not detected. The structural variants found in PSS were NAB2 intron 6-STAT6 intron 15 (rearranged gene including NAB2 e1/6- STAT6 e16/22, case 1), TPM3 intron 6-NTRK1 intron 8 (rearranged gene including TPM3 e1/6-NTRK1 e9/17, case 2), BCOR exon 14 with an unidentified partner (case 3), and a TP53 exon 1 rearrangement leading to predicted disruption of TP53 transcription (case 4). The structural variants found in STUMP were JAZF1 intron 3-SUZ12 exon 1 (rearranged gene including JAZF1 e1/3- SUZ12 e1/16, case 11), RGPD2 intron 1-FOXO1 intron 1, CLOCK exon 22-TP53 intron 1, and PALB2 exon 4-intergenic region of chromosome 2 (case 13). The TP53 and PALB2 fusions identified herein are expected to lead to inactivation of these genes rather than to productive and/or activating rearrangements. In case 13, PALB2 is the candidate oncogenic driver, possibly leading to homozygous recombination deficiency.
The diagram summarizes the results of DNA sequencing in 25 tumors of the specialized prostatic stroma. In case 11, synchronous STUMP and PSS components were differentially dissected and sequenced in parallel. Abbreviations: CNV copy number variant, NGS next-generation sequencing, STUMP stromal tumor of uncertain malignant potential, PSS prostatic stromal sarcoma. To simplify the figure, only chromosomes with arm-level or chromosome-level copy number changes are depicted.
One or more pathogenic somatic mutations were found in 15 cases (15/25, 60%; 15/21 successfully sequenced, 71%), including 8 PSS (8/11, 72%) and 7 STUMP (7/14, 50%). The only recurrently mutated genes were TP53 in 5 cases (5/25, 20%; 2/11, 18% PSS and 3/14, 21% STUMP) and ATRX in 2 cases [2/25, 8%; both PSS (2/11, 18%)]. In general, mutations were more frequent and numerous in cases without oncogenic rearrangements. The specific mutations present in each case are detailed in Table 3.
Copy number changes, including high-level amplifications and double copy deletions, were more frequent and numerous in cases without oncogenic gene rearrangements. Recurrent CNVs were −13q, −14q, −16q, and +8/+8q, each present in a small number of cases. Interestingly, case 6 had multiple arm-level and chromosome-level losses, with a near-haploid copy number. Another remarkable finding was the presence of focal, high-level co-amplification of MDM2, CDK4 and ERBB3 in case 19.
Case 11, a high-grade PSS associated with adjacent myxoid STUMP components, merits separate discussion. In this sample, the two components were dissected and sequenced in parallel. The STUMP component had TP53 and CHEK2 mutations, with the latter being likely germline based on the variant allele frequency. In addition, single copy loss of TP53 was also detected, suggesting that a subpopulation of the STUMP cells harbored homozygous TP53 inactivation. Chromosome-level and arm-level copy number changes were not identified in the STUMP (Fig. 7). The PSS component of case 11 had biallelic (homozygous) inactivation of both TP53 and CHEK2. Moreover, there were frequent copy number gains and losses and widespread loss of heterozygosity in a pattern consistent with whole-genome doubling. Specifically, analysis of the SNP profile of the PSS component demonstrated a 2:1 ratio of major-to-minor alleles in copy-neutral regions of the genome, most consistent with triploidy. The findings in case 11 propose a potential molecular correlate of biologic progression from STUMP to PSS, in which biallelic inactivation of TP53 and CHEK2 leads to cell cycle dysregulation in a subpopulation of STUMP cells prone to DNA damage, followed by genome endoreduplication events and widespread loss of heterozygosity that ultimately result in transformation to sarcoma.
Comparison of genome-wide copy number variant profiles demonstrates multiple regional, arm-level, and chromosome-level copy number changes as well as high-level amplifications in the PSS component, which are not seen in the STUMP component. Evaluation of genome-wide copy number and SNP profile of the PSS component show widespread loss of heterozygosity and findings consistent with triploidy. Copy number changes are expressed as log2 ratios. Abbreviations: STUMP stromal tumor of uncertain malignant potential, PSS prostatic stromal sarcoma.
RNA sequencing
RNA sequencing was performed on 17 cases (7 PSS, 10 STUMP) with remaining FFPE tissue available after DNA sequencing (cases 1, 3–5, 7–9, 13, 14, and 16–23, Table 3). Sequencing was successful in 15 cases (15/17, 88%) and failed in 2 cases (2/17, 12%). Gene fusions were found in 4 cases (4/15, 27%). The panel confirmed the presence of a NAB2-STAT6 fusion in case 1 (PSS) and demonstrated a BCOR-MAML3 fusion in case 3 (PSS). The latter event had been detected by DNA sequencing as a BCOR rearrangement with an unidentified partner. CIC fusion products were found in cases 14 and 22 (both STUMP), but they probably represented stochastic (i.e., random) events. This interpretation is supported by the frequent occurrence of low-confidence CIC fusion calls in our platform and by the absence of CIC rearrangements detected by OncoPanel and FISH (see below). Finally, multiple fusion events involving genes mapping to 12q15 were identified in case 19. As previously described [14], this finding is usually a surrogate marker for the presence of MDM2 and CDK4 amplification.
Fluorescence in situ hybridization (FISH)
Break apart FISH to investigate CIC rearrangement was performed in cases 14 and 22. The former failed hybridization despite multiple attempts and the latter was negative for CIC rearrangement.
Pathology re-review of cases with gene rearrangements/fusions
Cases with previously characterized gene fusions were re-reviewed to evaluate for morphologic features associated with each individual rearrangement.
Case 1 (NAB2-STAT6) was homogeneously hypercellular and consisted of patternless sheets of spindle and ovoid cells with round to ovoid nuclei, a scant amount of cytoplasm, and minimal nuclear atypia. There was a rich vascular network of prominent capillaries and arterioles with somewhat thick muscular walls that resembled spiral arteries of the endometrium. Foci with a short fascicular arrangement were noted and ectatic branching (“HPC-like”) vessels were not present.
Case 2 (TPM3-NTRK1) consisted of a homogeneously cellular proliferation of bland spindle cells arranged in short fascicles. Nuclear atypia was mild, but mitotic activity was prominent. There was a delicate collagenous stroma, but prominent collagenous bands and perivascular hyalinized collagen were not identified.
Case 3 (BCOR-MAML3) was a discrete, relatively well-circumscribed tumor nodule entirely confined to the prostate. Tumor borders were well demarcated, with only focal invasion of the adjacent prostatic stroma. The lesion consisted of highly cellular sheets of somewhat primitive-looking spindle and ovoid cells with round to ovoid nuclei, minimal nuclear atypia, and vaguely granular eosinophilic cytoplasm arranged in patternless fashion. Mitotic activity was prominent and tumor necrosis was present. A rich capillary network was noted, but myxoid stromal changes were not identified.
Case 12 (JAZF1-SUZ12) was hypercellular, with solid and partially tubular architecture resembling “sex-cord-like” elements. Lesional cells were small, spindle, and round, with a scant amount of amphophilic cytoplasm. Nuclear atypia was minimal, and mitotic activity was not identified (<1 per 10 hpf). Prominent capillaries and arterioles with conspicuous muscular walls reminiscent of the spiral arteries of the endometrium were easily identified.
In all, the only one of these four cases in which the presence of the specific gene fusion could have been suspected based on morphology was case 12. The remaining cases did not have distinctive morphologic features, and the underlying molecular alterations could not have been predicted based on their microscopic appearances.
Discussion
This study found that, from a molecular perspective, primary mesenchymal neoplasms of the prostate originally diagnosed as PSS and STUMP are highly heterogeneous. Except for a few mutations and CNVs present in small subsets of cases, genomic alterations were largely nonrecurrent. Unexpectedly, a subgroup of our cases seems to be driven by well-defined oncogenic gene fusions, including rearrangements seen in specific mesenchymal tumors of the uterus and soft tissues.
Historically, primary mesenchymal tumors of the prostate have been classified as STUMP or PSS based on their overall pathologic features, and both categories encompass neoplasms with a wide morphologic spectrum [1, 2]. Most patients present with lower urinary tract symptoms and/or hematuria [1, 7, 17], but many are diagnosed in asymptomatic men with abnormal digital rectal examination findings or elevated PSA levels [1]. STUMP commonly have an indolent clinical course without progression after non-radical resection (transurethral resection, prostatic enucleation) or during surveillance in patients that do not undergo ablative treatment after biopsy [1, 7, 17]. However, a subset of STUMP is associated with synchronous or metachronous PSS, sometimes with local recurrences and metastases resulting in disease-specific mortality [1, 7, 17]. Both low-grade and high-grade PSS, either pure or associated with STUMP, have potential for local progression and distant spread [1, 2, 7].
Data on the molecular characteristics and genetic drivers of these tumors are limited. Two prior studies of overlapping series have identified losses of chromosomes 13, 14, and 10 as the most common recurrent copy number events in the family of prostatic stromal neoplasia [8, 9]. However, recurrent cancer-relevant somatic mutations or gene fusions have not previously been identified [9]. The present study found that almost a quarter of cases (6/25, 24%; cases 4, 5, 11, and 14–16), including cases originally diagnosed as both PSS and STUMP, harbored pathogenic TP53 variants that likely resulted in functional inactivation of the gene. Moreover 2 cases (2/25, 8%) had truncating ATRX mutations and nonrecurrent pathogenic (i.e., cancer-relevant) mutations/indels were seen in 9 additional cases that lacked TP53 mutations, ATRX mutations and oncogenic gene fusions (9/25, 36%; cases 3, 6–9, 12 and 17–19). The mutations reported previously in prostatic stromal neoplasms were most likely non-pathogenic and did not involve the genes that were mutated in the present series [9].
An ever-expanding number of gene fusions are being described in a broad range of carcinomas and sarcomas. Whether gene rearrangements are definitional of specific tumor types is a somewhat debated issue, as many individual gene fusions have been identified in neoplasms with widely variable morphology, immunophenotype and biologic potential. However, a few rearrangements, such as NAB2-STAT6, are considered pathognomonic [18, 19] or show a very strong association with certain tumor types [20, 21]. In this study, driver fusions and/or rearrangements were detected in 4 cases (4/25, 16%; cases 1–3 and 12) by DNA and/or RNA sequencing. RNA sequencing also demonstrated CIC fusions in 2 STUMP (cases 14 and 22). These fusions probably represent stochastic events rather than true drivers, given the absence of CIC rearrangements by DNA sequencing in case 14 and by DNA sequencing plus FISH in case 20. In addition, rearrangements that likely lead to loss of function of PALB2 and TP53 were found in 2 cases and a novel RGPD2-FOXO1 rearrangement was found in 1 case. Although case 1 (PSS) had a striking microscopic resemblance to low-grade endometrial stromal sarcoma, it should probably be interpreted as a high-risk solitary fibrous tumor in light of the identification of a NAB2-STAT6 fusion [18, 19]. JAZF1-SUZ12 fusions have been found in a subgroup of low-grade endometrial stromal sarcomas [20, 21] and in endometrial stromal nodules [22], but their occurrence in sarcomas presenting in male patients is exceptionally rare [23]. In case 12, the overall microscopic features and the presence of a JAZF1-SUZ12 fusion are highly suggestive of low-grade endometrial stromal sarcoma, and probably warrant a diagnosis of “low-grade endometrial sarcoma-like” sarcoma. TPM3-NTRK1 has been demonstrated in uterine sarcomas [24, 25], in soft tissue tumors with lipofibromatosis-like morphology and a “neural” immunophenotype [26], as well as in carcinomas [27, 28]. According to current diagnostic practices, case 2 can be considered an intermediate grade sarcoma with NTRK1 rearrangement [29]. BCOR fusions, including BCOR-MAML3, have been identified in a heterogeneous group of sarcomas that includes a subset of tumors with small round blue cell morphology [30,31,32]. Similar to the prior tumor, case 3 can be classified as an intermediate grade sarcoma with BCOR rearrangement. A classification based on the underlying rearrangement is more problematic in case 13, since the RGPD2-FOXO1 rearrangement has not previously been reported, to the best of our knowledge. Novel non-canonical partners have been identified in sarcomas with FOXO1 rearrangments [33] and this RGPD2-FOXO1 fusion was in-frame. However, the absence of an RGPD2-FOXO1 fusion product demonstrated by RNA sequencing indicates the presence of a non-productive rearrangement.
A subset of PSS, including 2 cases with concurrent ATRX mutations, showed marked copy number instability (cases 4–8). The present series showed low-frequency recurrent copy number losses involving large areas of chromosomes 13, 14, and 16. Recurrent gains of chromosome 8/8p were also identified in three cases. Concurrent pathogenic mutations were present in all cases with copy number losses of chromosomes 13 and 14. A rather interesting finding was the presence of high-level MDM2-CDK4-ERBB3 co-amplification in a STUMP (case 19). In this sample (simple prostatectomy), the lesion had bland spindle cell morphology without prominent mitotic activity. Although MDM2-CDK4 co-amplification is considered a hallmark of atypical lipomatous tumor/well-differentiated liposarcoma and dedifferentiated liposarcoma, it is by no means specific and has been also described in multiple sarcomas, carcinomas, and sex-cord stromal tumors [34,35,36]. Interestingly, case 19 also harbored a POT1 mutation, which might be associated causally with the alterations seen in chromosome 12. POT1 is a member of the shelterin complex that binds to single-stranded DNA overhangs to stabilize the telomeres, and POT1 mutations have been associated with structural chromosomal abnormalities [37].
An important finding of this study was that a subset of primary prostatic mesenchymal neoplasms originally considered within the spectrum of specialized prostatic stromal neoplasia could be reclassified using molecular analyses (Table 4). Specifically, 4 cases (4/25, 16%) harbored rearrangements and/or gene fusions that were useful for their categorization, namely: NAB2-STAT6, TMP3-NTRK1, JAZF1-SUZ12, and BCOR-MAML3. Another relevant finding is that most of our cases (19/22 cases that underwent successful DNA and/or RNA sequencing, 86%) had pathogenic genetic variants, some of which might be relevant for treatment purposes. In this regard, genetic variants might identify cases amenable to treatment with agents that target specific fusion proteins (e.g., case 2) [38] or molecular pathways (e.g., case 8) [39, 40]. Moreover, the molecular findings can also guide treatment by properly classifying the tumors (e.g., case 1 and 12) or by suggesting susceptibility to specific agents (e.g., cases 4, 5, 14, and 16) [41,42,43,44] in these rare lesions that lack a standardized therapeutic approach.
Classification of mesenchymal tumors of the prostate as benign or malignant is sometimes difficult, justifying the existence of STUMP as a diagnostic category for tumors of uncertain biologic potential. Although prior series have described histologic features that can help distinguish between PSS and STUMP [2], there are no objective and validated diagnostic criteria, mostly due to the rarity of these tumors. It is therefore likely that neoplasms historically classified as STUMP comprise a broad range of lesions, including non-neoplastic stromal growths (e.g., florid stromal hyperplasia), indolent neoplasms, and low-grade sarcomas. This hypothesis is supported by the results of the molecular analyses performed herein. For instance, cases originally classified as STUMP that harbor biallelic TP53 inactivation (cases 14 and 16) or a known driver fusion (case 12) are most likely sarcomas. In case 15, a pathogenic likely heterozygous TP53 mutation was identified by DNA sequencing. This tumor progressed with intraabdominal metastases that had the same morphology but higher histologic grade than the primary lesion. It is likely that biallelic loss of TP53 due to a “second hit” parallels the development of aggressive biology in some of these tumors that would otherwise be classified as “of uncertain malignant potential” (STUMP). Therefore, the presence of even a heterozygous pathogenic TP53 mutation should be considered an indicator of malignant potential. This is nicely illustrated by case 11, in which a bland-appearing component of the tumor with subclonal biallelic TP53 inactivation is associated with an adjacent high-grade sarcoma that harbors widepread biallelic TP53 inactivation as well as homozygous CHEK2 inactivation and numerous copy number changes. Moreover, based on data from soft tissue tumors with high-level MDM2 and CDK4 amplification [45,46,47,48], case 19 probably has malignant potential. The above findings demonstrate that molecular evaluation may be useful to better understand the underlying biology and classify these tumors as benign or malignant.
The results of this study suggest that, from a molecular perspective, so-called tumors of specialized prostatic stroma (STUMP and PSS) are highly heterogeneous and may perhaps not represent a discrete entity. Of note, this study includes a somewhat larger proportion of sarcomas than prior studies [8, 9]. However, unique cancer-relevant genetic variants were also found in most STUMP. The data presented herein demonstrates that tumors historically regarded as specialized prostatic stromal lineage probably comprise a heterogeneous group of mesenchymal neoplasms that also occur in other anatomic sites. Being diagnoses of exclusion, STUMP and PSS are likely repositories for diverse neoplasms that cannot be further classified with the techniques available at the time of diagnosis. This is supported by a subset of tumors from this retrospective series that could be reclassified by NGS, and by the presence of largely nonrecurrent cancer-relevant genetic variants in most cases. The latter finding suggests that these sundry lesions have diverse genetic backgrounds and, most likely, disparate molecular drivers. Based on the results presented herein, there is no definite evidence to support the existence of tumors of the specialized prostatic stroma as a distinct biologic entity. Therefore, a reconsideration of the current diagnostic terminology might be appropriate until further confirmatory studies are performed. In this regard, for cases that cannot currently be classified, “low/intermediate/high-grade sarcoma, unclassified” and “mesenchymal neoplasm of uncertain biologic potential” could perhaps be used instead of PSS and STUMP, respectively. From a practical diagnostic work-up perspective, STAT6 screening IHC in this setting seems warranted, as SFT is well documented in the prostate and many cases have unusual or high-grade features [49, 50].
The presence of chromosome 13 and 14 losses and the absence of cancer-relevant somatic mutations and gene rearrangements [9] have been considered characteristic molecular features of tumors of purported specialized prostatic stromal origin. However, our analysis demonstrates that most mesenchymal tumors of the prostate harbor pathogenic mutations and/or gene rearrangements/fusions, including cases with concurrent loss of large areas of chromosomes 13 and 14 (Fig. 7). This discrepancy can be explained in part by the use of different platforms for molecular profiling [9]. Whole-exome sequencing generally has a more limited depth of coverage than targeted panels and has limited ability to detect gene rearrangements. Therefore, the broad coverage achieved with whole-exome sequencing comes at the cost of limited sensitivity for SNVs, indels, and structural variants. In the present study, mutations, rearrangements and fusions were investigated with well-established clinically validated platforms for targeted DNA and RNA sequencing. In addition, the last few years have seen a surprisingly rapid improvement of the bioinformatic pipelines, resulting in improved sensitivity and specificity for variant calling.
This study has limitations that need to be addressed. For instance, the number of cases analyzed is relatively small, albeit comparable to the size of prior series with molecular analyses[8, 9]. Also, benign tissue for assessment of germline status was not available; nonetheless, sequencing metrics such as the variant allele fraction were used to infer the somatic nature of mutations and indels [51]. Despite the above-mentioned shortcomings, this study provides a comprehensive molecular profiling of a multi-institutional series of mesenchymal tumors of purported specialized prostatic stromal origin using both DNA and RNA sequencing.
In summary, the present study demonstrated that primary mesenchymal neoplasms of the prostate are molecularly heterogeneous and include cases that can be classified more specifically according to the underlying genetic alterations. Also, a subset of cases has potentially actionable molecular findings. Importantly, these results suggest that mesenchymal tumors of the prostate do not represent a distinct biologic entity, which may justify a reconsideration of the diagnostic terminology.
Data availability
The data generated during the current study are available from the corresponding author on reasonable request.
References
Herawi M, Epstein JI. Specialized stromal tumors of the prostate: a clinicopathologic study of 50 cases. Am J Surg Pathol. 2006;30:694–704.
Gaudin PB, Rosai J, Epstein JI. Sarcomas and related proliferative lesions of specialized prostatic stroma: a clinicopathologic study of 22 cases. Am J Surg Pathol. 1998;22:148–62.
Henricks WH, Chu YC, Goldblum JR, Weiss SW. Dedifferentiated liposarcoma: a clinicopathological analysis of 155 cases with a proposal for an expanded definition of dedifferentiation. Am J Surg Pathol. 1997;21:271–81.
Chen E, Fletcher CDM. Cellular angiofibroma with atypia or sarcomatous transformation: clinicopathologic analysis of 13 cases. Am J Surg Pathol. 2010;34:707–14.
Mosquera J-M, Fletcher CDM. Expanding the spectrum of malignant progression in solitary fibrous tumors: a study of 8 cases with a discrete anaplastic component-is this dedifferentiated SFT? Am J Surg Pathol. 2009;33:1314–21.
Antonescu CR, Romeo S, Zhang L, Nafa K, Hornick JL, Petur Nielsen G, et al. Dedifferentiation in gastrointestinal stromal tumor to an anaplastic KIT-negative phenotype: a diagnostic pitfall: morphologic and molecular characterization of 8 cases occurring either de novo or after imatinib therapy. Am J Surg Pathol. 2013;37:385–92.
Bostwick DG, Hossain D, Qian J, Neumann RM, Yang P, Young RH, et al. Phyllodes tumor of the prostate: long-term followup study of 23 cases. J Urol. 2004;172:894–9.
Pan C-C, Epstein JI. Common chromosomal aberrations detected by array comparative genomic hybridization in specialized stromal tumors of the prostate. Mod Pathol. 2013;26:1536–43.
Pan C-C, Tsuzuki T, Morii E, Fushimi H, Chih-Hsueh Chen P, Epstein JI. Whole-exome sequencing demonstrates recurrent somatic copy number alterations and sporadic mutations in specialized stromal tumors of the prostate. Hum Pathol. 2018;76:9–16.
Garcia EP, Minkovsky A, Jia Y, Ducar MD, Shivdasani P, Gong X, et al. Validation of OncoPanel: a targeted next-generation sequencing assay for the detection of somatic variants in cancer. Arch Pathol Lab Med. 2017;141:751–8.
Sholl LM, Do K, Shivdasani P, Cerami E, Dubuc AM, Kuo FC, et al. Institutional implementation of clinical tumor profiling on an unselected cancer population. JCI Insight. 2016;1:e87062.
Abo RP, Ducar M, Garcia EP, Thorner AR, Rojas-Rudilla V, Lin L, et al. BreaKmer: detection of structural variation in targeted massively parallel sequencing data using kmers. Nucleic Acids Res. 2015;43:e19.
Papke DJ, Nowak JA, Yurgelun MB, Frieden A, Srivastava A, Lindeman NI, et al. Validation of a targeted next-generation sequencing approach to detect mismatch repair deficiency in colorectal adenocarcinoma. Mod Pathol. 2018;31:1882–90.
Dickson BC, Swanson D. Targeted RNA sequencing: a routine ancillary technique in the diagnosis of bone and soft tissue neoplasms. Genes Chromosomes Cancer. 2019;58:75–87.
Liu S, Tsai W-H, Ding Y, Chen R, Fang Z, Huo Z, et al. Comprehensive evaluation of fusion transcript detection algorithms and a meta-caller to combine top performing methods in paired-end RNA-seq data. Nucleic Acids Res. 2016;44:e47.
Chen X, Schulz-Trieglaff O, Shaw R, Barnes B, Schlesinger F, Källberg M, et al. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics. 2016;32:1220–2.
Shen Q, Zhou Z, Liu Z, Hu S, Lin Z, Li S, et al. Clinical and pathological features of prostatic stromal tumor of uncertain malignant potential: a retrospective study of 23 Chinese cases. Urol Int. 2021;105:206–14.
Mohajeri A, Tayebwa J, Collin A, Nilsson J, Magnusson L, von Steyern FV, et al. Comprehensive genetic analysis identifies a pathognomonic NAB2/STAT6 fusion gene, nonrandom secondary genomic imbalances, and a characteristic gene expression profile in solitary fibrous tumor. Genes Chromosomes Cancer. 2013;52:873–86.
Robinson DR, Wu Y-M, Kalyana-Sundaram S, Cao X, Lonigro RJ, Sung Y-S, et al. Identification of recurrent NAB2-STAT6 gene fusions in solitary fibrous tumor by integrative sequencing. Nat Genet. 2013;45:180–5.
Koontz JI, Soreng AL, Nucci M, Kuo FC, Pauwels P, van Den Berghe H, et al. Frequent fusion of the JAZF1 and JJAZ1 genes in endometrial stromal tumors. Proc Natl Acad Sci USA. 2001;98:6348–53.
Huang H-Y, Ladanyi M, Soslow RA. Molecular detection of JAZF1-JJAZ1 gene fusion in endometrial stromal neoplasms with classic and variant histology: evidence for genetic heterogeneity. Am J Surg Pathol. 2004;28:224–32.
Micci F, Heim S, Panagopoulos I. Molecular pathogenesis and prognostication of “low-grade” and “high-grade” endometrial stromal sarcoma. Genes Chromosomes Cancer. 2021;60:160–7.
Agaimy A, Moskalev EA, Weisser W, Bach T, Haller F, Hartmann A. Low-grade endometrioid stromal sarcoma of the paratestis: a novel report with molecular confirmation of JAZF1/SUZ12 translocation. Am J Surg Pathol. 2018;42:695–700.
Chiang S, Cotzia P, Hyman DM, Drilon A, Tap WD, Zhang L, et al. NTRK fusions define a novel uterine sarcoma subtype with features of fibrosarcoma. Am J Surg Pathol. 2018;42:791–8.
Croce S, Hostein I, Longacre TA, Mills AM, Pérot G, Devouassoux-Shisheboran M, et al. Uterine and vaginal sarcomas resembling fibrosarcoma: a clinicopathological and molecular analysis of 13 cases showing common NTRK-rearrangements and the description of a COL1A1-PDGFB fusion novel to uterine neoplasms. Mod Pathol. 2019;32:1008–22.
Agaram NP, Zhang L, Sung Y-S, Chen C-L, Chung CT, Antonescu CR, et al. Recurrent NTRK1 gene fusions define a novel subset of locally aggressive lipofibromatosis-like neural tumors. Am J Surg Pathol. 2016;40:1407–16.
Lasota J, Chłopek M, Lamoureux J, Christiansen J, Kowalik A, Wasąg B, et al. Colonic adenocarcinomas harboring NTRK fusion genes: a clinicopathologic and molecular genetic study of 16 cases and review of the literature. Am J Surg Pathol. 2020;44:162–73.
Xia H, Xue X, Ding H, Ou Q, Wu X, Nagasaka M, et al. Evidence of NTRK1 fusion as resistance mechanism to EGFR TKI in EGFR+ NSCLC: results from a large-scale survey of NTRK1 fusions in chinese patients with lung cancer. Clin Lung Cancer. 2020;21:247–54.
Suurmeijer AJH, Dickson BC, Swanson D, Zhang L, Sung Y-S, Cotzia P, et al. A novel group of spindle cell tumors defined by S100 and CD34 co-expression shows recurrent fusions involving RAF1, BRAF, and NTRK1/2 genes. Genes Chromosomes Cancer. 2018;57:611–21.
Specht K, Zhang L, Sung Y-S, Nucci M, Dry S, Vaiyapuri S, et al. Novel BCOR-MAML3 and ZC3H7B-BCOR gene fusions in undifferentiated small blue round cell sarcomas. Am J Surg Pathol. 2016;40:433–42.
Yoshida A, Arai Y, Hama N, Chikuta H, Bando Y, Nakano S, et al. Expanding the clinicopathologic and molecular spectrum of BCOR-associated sarcomas in adults. Histopathology. 2020;76:509–20.
Kao Y-C, Owosho AA, Sung Y-S, Zhang L, Fujisawa Y, Lee J-C, et al. BCOR-CCNB3 fusion positive sarcomas: a clinicopathologic and molecular analysis of 36 cases with comparison to morphologic spectrum and clinical behavior of other round cell sarcomas. Am J Surg Pathol. 2018;42:604–15.
Liu J, Guzman MA, Pezanowski D, Patel D, Hauptman J, Keisling M, et al. FOXO1-FGFR1 fusion and amplification in a solid variant of alveolar rhabdomyosarcoma. Mod Pathol. 2011;24:1327–35.
Dembla V, Somaiah N, Barata P, Hess K, Fu S, Janku F, et al. Prevalence of MDM2 amplification and coalterations in 523 advanced cancer patients in the MD Anderson phase 1 clinic. Oncotarget. 2018;9:33232–43.
Necchi A, Bratslavsky G, Shapiro O, Elvin JA, Vergilio J-A, Killian JK, et al. Genomic features of metastatic testicular sex cord stromal tumors. Eur Urol Focus. 2019;5:748–55.
Zhao J, Roth J, Bode-Lesniewska B, Pfaltz M, Heitz PU, Komminoth P. Combined comparative genomic hybridization and genomic microarray for detection of gene amplifications in pulmonary artery intimal sarcomas and adrenocortical tumors. Genes Chromosomes Cancer. 2002;34:48–57.
Chang S. Cancer chromosomes going to POT1. Nat Genet. 2013;45:473–5.
Assi T, Rassy E, Nassereddine H, Farhat F, El Karak F, Kattan J, et al. TRK inhibition in soft tissue sarcomas: a comprehensive review. Semin Oncol. 2020;47:73–84.
Dickson MA, Schwartz GK, Antonescu CR, Kwiatkowski DJ, Malinowska IA. Extrarenal perivascular epithelioid cell tumors (PEComas) respond to mTOR inhibition: clinical and molecular correlates. Int J Cancer. 2013;132:1711–7.
Kwiatkowski DJ, Choueiri TK, Fay AP, Rini BI, Thorner AR, de Velasco G, et al. Mutations in TSC1, TSC2, and MTOR are associated with response to rapalogs in patients with metastatic renal cell carcinoma. Clin Cancer Res. 2016;22:2445–52.
McCabe N, Turner NC, Lord CJ, Kluzek K, Bialkowska A, Swift S, et al. Deficiency in the repair of DNA damage by homologous recombination and sensitivity to poly(ADP-ribose) polymerase inhibition. Cancer Res. 2006;66:8109–15.
Koschmann C, Calinescu A-A, Nunez FJ, Mackay A, Fazal-Salom J, Thomas D, et al. ATRX loss promotes tumor growth and impairs nonhomologous end joining DNA repair in glioma. Sci Transl Med. 2016;8:328ra28.
Villarroel MC, Rajeshkumar NV, Garrido-Laguna I, De Jesus-Acosta A, Jones S, Maitra A, et al. Personalizing cancer treatment in the age of global genomic analyses: PALB2 gene mutations and the response to DNA damaging agents in pancreatic cancer. Mol Cancer Ther. 2011;10:3–8.
George SL, Lorenzi F, King D, Hartlieb S, Campbell J, Pemberton H, et al. Therapeutic vulnerabilities in the DNA damage response for the treatment of ATRX mutant neuroblastoma. EBioMedicine. 2020;59:102971.
Dewaele B, Floris G, Finalet-Ferreiro J, Fletcher CD, Coindre J-M, Guillou L, et al. Coactivated platelet-derived growth factor receptor {alpha} and epidermal growth factor receptor are potential therapeutic targets in intimal sarcoma. Cancer Res. 2010;70:7304–14.
Sirvent N, Coindre J-M, Maire G, Hostein I, Keslair F, Guillou L. et al. Detection of MDM2-CDK4 amplification by fluorescence in situ hybridization in 200 paraffin-embedded tumor samples: utility in diagnosing adipocytic lesions and comparison with immunohistochemistry and real-time PCR. Am J Surg Pathol. 2007;31:1476–89.
Neuville A, Collin F, Bruneval P, Parrens M, Thivolet F, Gomez-Brouchet A, et al. Intimal sarcoma is the most frequent primary cardiac sarcoma: clinicopathologic and molecular retrospective analysis of 100 primary cardiac sarcomas. Am J Surg Pathol. 2014;38:461–9.
Pedeutour F, Suijkerbuijk RF, Forus A, Van Gaal J, Van de Klundert W, Coindre JM, et al. Complex composition and co-amplification of SAS and MDM2 in ring and giant rod marker chromosomes in well-differentiated liposarcoma. Genes Chromosomes Cancer. 1994;10:85–94.
Guner G, Bishop JA, Bezerra SM, Taheri D, Zahavi DJ, Mendoza Rodriguez MA, et al. The utility of STAT6 and ALDH1 expression in the differential diagnosis of solitary fibrous tumor versus prostate-specific stromal neoplasms. Hum Pathol. 2016;54:184–8.
Bakhshwin A, Berry RS, Cox RM, Li R, Reynolds JP, Rubin BP, et al. Malignant solitary fibrous tumour of the prostate: four cases emphasising significant histological and immunophenotypical overlap with sarcomatoid carcinoma. Pathology. 2020;52:643–8.
Li MM, Datto M, Duncavage EJ, Kulkarni S, Lindeman NI, Royet S, et al. Standards and guidelines for the interpretation and reporting of sequence variants in cancer: a joint consensus recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J Mol Diagn. 2017;19:4–23.
Acknowledgements
We would like to acknowledge the following colleagues for their important contribution to this study: Dr. Michelle S. Hirsch, MD, Ph.D.; Department of Pathology, Brigham and Women’s Hospital, Boston, MA. Dr. Roanh Le Dinh, MD; Center for Research and Early Detection of Cancer, Ha Noi, Vietnam. Dr. Neil Lindeman, MD; Center for Advanced Molecular Diagnostics (CAMD), Department of Pathology, Brigham and Women’s Hospital, Boston, MA.
Funding
The cost of the RNA sequencing assays was covered by the Panov 2 Research Fund (BCD)
Author information
Authors and Affiliations
Contributions
Concept, design and coordination: AMA. Analysis and interpretation of sequencing data: LMS, BCD, AM-E, AMD. Evaluation of Fluorescence In Situ Hybridization (FISH) results: PDC. Review of cases, interpretation of results and correlation of histologic and molecular data: AMA and CDF. Manuscript drafting and figure design: AMAcosta. Contribution of cases: CDF, JKM, JBG, MRP. Manuscript review and editing: All authors. Intellectual input/contributions: All authors
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Acosta, A.M., Sholl, L.M., Dickson, B.C. et al. Re-evaluating tumors of purported specialized prostatic stromal origin reveals molecular heterogeneity, including non-recurring gene fusions characteristic of uterine and soft tissue sarcoma subtypes. Mod Pathol 34, 1763–1779 (2021). https://doi.org/10.1038/s41379-021-00818-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41379-021-00818-6
This article is cited by
-
ARID1A deficient undifferentiated spindle cell and rhabdoid sarcoma of the prostate: report of a unique case with emphasis on diagnostic implications
Diagnostic Pathology (2022)
-
Adult NTRK-rearranged spindle cell neoplasms of the viscera: with an emphasis on rare locations and heterologous elements
Modern Pathology (2022)
-
Reexamining the molecular findings in specialized stromal tumors of the prostate
Modern Pathology (2021)
-
In Response to “Reexamining the molecular findings in specialized stromal tumors of the prostate”
Modern Pathology (2021)