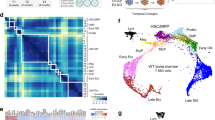
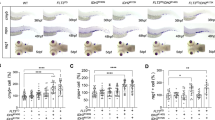
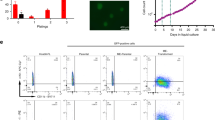
Abstract
CRISPR-mediated simultaneous targeting of candidate tumor suppressor genes in Xenopus tropicalis allows fast functional assessment of co-driver genes for various solid tumors. Genotyping of tumors that emerge in the mosaic mutant animals rapidly exposes the gene mutations under positive selection for tumor establishment. However, applying this simple approach to the blood lineage has not been attempted. Multiple hematologic malignancies have mutations in EZH2, encoding the catalytic subunit of the Polycomb Repressive Complex 2. Interestingly, EZH2 can act as an oncogene or a tumor suppressor, depending on cellular context and disease stage. We show here that mosaic CRISPR/Cas9 mediated ezh2 disruption in the blood lineage resulted in early and penetrant acute myeloid leukemia (AML) induction. While animals were co-targeted with an sgRNA that induces notch1 gain-of-function mutations, sequencing of leukemias revealed positive selection towards biallelic ezh2 mutations regardless of notch1 mutational status. Co-targeting dnm2, recurrently mutated in T/ETP-ALL, induced a switch from myeloid towards acute T-cell leukemia. Both myeloid and T-cell leukemias engrafted in immunocompromised hosts. These data underline the potential of Xenopus tropicalis for modeling human leukemia, where mosaic gene disruption, combined with deep amplicon sequencing of the targeted genomic regions, can rapidly and efficiently expose co-operating driver gene mutations.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All data supporting the findings of this study are available within the article and its supplementary information and from the corresponding author upon reasonable request.
References
Liu Y, Easton J, Shao Y, Maciaszek J, Wang Z, Wilkinson MR, et al. The genomic landscape of pediatric and young adult T-lineage acute lymphoblastic leukemia. Nat Genet. 2017;49:1211–8.
George J, Lim JS, Jang SJ, Cun Y, Ozretia L, Kong G, et al. Comprehensive genomic profiles of small cell lung cancer. Nature. 2015;524:47–53.
McDonald SL, Silver A. The opposing roles of Wnt-5a in cancer. Br J Cancer. 2009;101:209–14.
Yap LF, Lee D, Khairuddin ANM, Pairan MF, Puspita B, Siar CH, et al. The opposing roles of NOTCH signalling in head and neck cancer: a mini review. Oral Dis. 2015;21:850–7.
Van Nieuwenhuysen T, Naert T, Tran HT, Van Imschoot G, Geurs S, Sanders E, et al. TALEN-mediated apc mutation in Xenopus tropicalis phenocopies familial adenomatous polyposis. Oncoscience. 2015;2:555–66.
Naert T, Dimitrakopoulou D, Tulkens D, Demuynck S, Carron M, Noelanders R, et al. RBL1 (p107) functions as tumor suppressor in glioblastoma and small-cell pancreatic neuroendocrine carcinoma in Xenopus tropicalis. Oncogene. 2020;39:2692–706.
Naert T, Tulkens D, Van Nieuwenhuysen T, Przybyl J, Demuynck S, Van de Rijn M, et al. CRISPR-SID: Identifying EZH2 as a druggable target for desmoid tumors via in vivo dependency mapping. Proc Natl Acad Sci USA. 2021;118:e2115116118.
Naert T, Colpaert R, Van Nieuwenhuysen T, Dimitrakopoulou D, Leoen J, Haustraete J, et al. CRISPR/Cas9 mediated knockout of rb1 and rbl1 leads to rapid and penetrant retinoblastoma development in Xenopus tropicalis. Sci Rep. 2016;6:1–10.
Tulkens D, Vleminckx K. Studying tumor formation and regulation in xenopus. CRC Press: Boca Raton, 2022, p. 301–11.
Comet I, Riising EM, Leblanc B, Helin K. Maintaining cell identity: PRC2-mediated regulation of transcription and cancer. Nat Rev Cancer. 2016;16:803–10.
Kim KH, Roberts CWM. Targeting EZH2 in cancer. Nat Med. 2016;22:128–34.
Chang CJ, Hung MC. The role of EZH2 in tumour progression. Br J Cancer. 2011;106:243–7.
Simon JA, Lange CA. Roles of the EZH2 histone methyltransferase in cancer epigenetics. Mutat Res Mol Mech Mutagen. 2008;647:21–29.
Safaei S, Baradaran B, Hagh MF, Alivand MR, Talebi M, Gharibi T, et al. Double sword role of EZH2 in leukemia. Biomed Pharmacother. 2018;98:626–35.
Ntziachristos P, Tsirigos A, Welstead GG, Trimarchi T, Bakogianni S, Xu L, et al. Contrasting roles of histone 3 lysine 27 demethylases in acute lymphoblastic leukaemia. Nature. 2014;514:513–7.
Neumann M, Heesch S, Schlee C, Schwartz S, Gökbuget N, Hoelzer D, et al. Whole-exome sequencing in adult ETP-ALL reveals a high rate of DNMT3A mutations. Blood. 2013;121:4749–52.
Van Vlierberghe P, Ambesi-Impiombato A, De Keersmaecker K, Hadler M, Paietta E, Tallman MS, et al. Prognostic relevance of integrated genetic profiling in adult T-cell acute lymphoblastic leukemia. Blood. 2013;122:74–82.
Ge Z, Li M, Zhao G, Xiao L, Gu Y, Zhou X, et al. Novel dynamin 2 mutations in adult T-cell acute lymphoblastic leukemia. Oncol Lett. 2016;12:2746–51.
Moreno-Mateos MA, Vejnar CE, Beaudoin J-D, Fernandez JP, Mis EK, Khokha MK, et al. CRISPRscan: designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat Methods. 2015;12:982–8.
Boel A, Steyaert W, De Rocker N, Menten B, Callewaert B, De Paepe A, et al. BATCH-GE: Batch analysis of Next-Generation Sequencing data for genome editing assessment. Sci Rep. 2016;6:30330.
Naert T, Tulkens D, Edwards NA, Carron M, Shaidani N-I, Wlizla M, et al. Maximizing CRISPR/Cas9 phenotype penetrance applying predictive modeling of editing outcomes in Xenopus and zebrafish embryos. Sci Rep. 2020;10:1–12.
Maxham LA, Forzán MJ, Hogan NS, Vanderstichel RV, Gilroy CV. Hematologic reference intervals for Xenopus tropicalis with partial use of automatic counting methods and reliability of long-term stored samples. Vet Clin Pathol. 2016;45:291–9.
Natt MP, Herrick CA. A new blood diluent for counting the erythrocytes and leucocytes of the chicken. Poult Sci. 1952;31:735–8.
Rogers S, Wells R, Rechsteiner M. Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science. 1986;234:364–8.
Sanchez-Martin M, Ferrando A. The NOTCH1-MYC highway toward T-cell acute lymphoblastic leukemia. Blood. 2017;129:1124–33.
Ciau-Uitz A, Patient R. The embryonic origins and genetic programming of emerging haematopoietic stem cells. FEBS Lett. 2016;590:4002–15.
Robert J, Ohta Y. Comparative and developmental study of the immune system in Xenopus. Dev Dyn. 2009;238:1249–70.
Tulkens D, Dimitrakopoulou D, Boelens M, Van Nieuwenhuysen T, Demuynck S, Toussaint W, et al. Engraftment of Allotransplanted Tumor Cells in Adult rag2 Mutant Xenopus tropicalis. Cancers. 2022;14:4560.
Zhang J, Ding L, Holmfeldt L, Wu G, Heatley SL, Payne-Turner D, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481:157–63.
Curtis DJ, Brown FC, Collett M, Lucas SE, Saw J, Robinson PJ, et al. Heterozygous mutation of dynamin 2 expands the pool of IL-7 responsive leukemic stem cells in T-cell acute lymphoblastic leukemia. Blood. 2013;122:613–613.
Tremblay CS, Brown FC, Collett M, Saw J, Chiu SK, Sonderegger SE, et al. Loss-of-function mutations of Dynamin 2 promote T-ALL by enhancing IL-7 signalling. Leukemia. 2016;30:1993–2001.
Bankhead P, Loughrey MB, Fernández JA, Dombrowski Y, McArt DG, Dunne PD, et al. QuPath: open source software for digital pathology image analysis. Sci Rep. 2017;7:1–7.
Oliveira ML, Akkapeddi P, Ribeiro D, Melão A, Barata JT. IL-7R-mediated signaling in T-cell acute lymphoblastic leukemia: an update. Adv Biol Regul. 2019;71:88–96.
Naert T, Colpaert R, Van Nieuwenhuysen T, Dimitrakopoulou D, Leoen J, Haustraete J, et al. CRISPR/Cas9 mediated knockout of rb1 and rbl1 leads to rapid and penetrant retinoblastoma development in Xenopus tropicalis. Sci Rep. 2016;6:35264.
Mann KM, Newberg JY, Black MA, Jones DJ, Amaya-Manzanares F, Guzman-Rojas L, et al. Analyzing tumor heterogeneity and driver genes in single myeloid leukemia cells with SBCapSeq. Nat Biotechnol. 2016;34:962–72.
Shen L, Shi Q, Wang W. Double agents: genes with both oncogenic and tumor-suppressor functions. Oncogenesis. 2018;7:25.
Schäfer V, Ernst J, Rinke J, Winkelmann N, Beck JF, Hochhaus A, et al. EZH2 mutations and promoter hypermethylation in childhood acute lymphoblastic leukemia. J Cancer Res Clin Oncol. 2016;142:1641–50.
Harikrishnan KN, Bassal S, Tikellis C, El-Osta A. Expression analysis of the epigenetic methyltransferases and methyl-CpG binding protein families in the normal B-cell and B-cell chronic lymphocytic leukemia (CLL). Cancer Biol Ther. 2004;3:989–94.
Nishioka C, Ikezoe T, Yang J, Yokoyama A. BCR/ABL increases EZH2 levels which regulates XIAP expression via miRNA-219 in chronic myeloid leukemia cells. Leuk Res. 2016;45:24–32.
Van Der Meulen J, Van Roy N, Van Vlierberghe P, Speleman F. The epigenetic landscape of T-cell acute lymphoblastic leukemia. Int J Biochem Cell Biol. 2014;53:547–57.
Ernst T, Chase AJ, Score J, Hidalgo-Curtis CE, Bryant C, Jones AV, et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat Genet. 2010;42:722–6.
Xu F, Li X, Wu L, Zhang Q, Yang R, Yang Y, et al. Overexpression of the EZH2, RING1 and BMI1 genes is common in myelodysplastic syndromes: relation to adverse epigenetic alteration and poor prognostic scoring. Ann Hematol. 2011;90:643–53.
Göllner S, Oellerich T, Agrawal-Singh S, Schenk T, Klein HU, Rohde C, et al. Loss of the histone methyltransferase EZH2 induces resistance to multiple drugs in acute myeloid leukemia. Nat Med. 2017;23:69–78.
Basheer F, Giotopoulos G, Meduri E, Yun H, Mazan M, Sasca D, et al. Contrasting requirements during disease evolution identify EZH2 as a therapeutic target in AML. J Exp Med. 2019;216:966–81.
Vardiman JW, Harris NL, Brunning RD. The World Health Organization (WHO) classification of the myeloid neoplasms. Blood. 2002;100:2292–302.
O’Carroll D, Erhardt S, Pagani M, Barton SC, Surani MA, Jenuwein T. The polycomb-group gene Ezh2 is required for early mouse development. Mol Cell Biol. 2001;21:4330–6.
Felice MS, Hammermuller E, De Dávila MT, Ciocca ME, Fraquelli LE, Lorusso AM, et al. Acute lymphoblastic leukemia presenting as acute hepatic failure in childhood. Leuk Lymphoma. 2009;38:633–7.
Heincelman M, Karakala N, Rockey DC. Acute lymphoblastic leukemia in a young adult presenting as hepatitis and acute kidney injury. J Investig Med High Impact Case Rep. 2016;4:1–4.
Donnelly WH, Miale TD. Respiratory distress secondary to pulmonary leukemia. Am J Dis Child. 1981;135:716–8.
Guermazi A, Feger C, Rousselot P, Merad M, Benchaib N, Bourrier P, et al. Granulocytic sarcoma (chloroma): imaging findings in adults and children. Am J Roentgenol. 2002;178:319–25.
Umer I, Umer I, Burki F, Ahmed Y. Bilateral leukemic pulmonary infiltrate: a rare manifestation of acute myeloid leukemia. J Med Cases. 2014;5:137–9.
Potenza L, Luppi M, Morselli M, Tonelli S, D’Apollo N, Facchini L, et al. Leukaemic pulmonary infiltrates in adult acute myeloid leukaemia: a high-resolution computerized tomography study. Br J Haematol. 2003;120:1058–61.
Acknowledgements
DT was funded by a Ph.D. fellowship from the Research Foundation—Flanders (FWO-Vlaanderen, 3F021818). Research in the authors’ laboratory is supported by the Research Foundation—Flanders (FWO-Vlaanderen) (grants 3G0A6922 and 3G0D8716) and by the Concerted Research Actions from Ghent University (01G01115). Further support was obtained by the Hercules Foundation, Flanders (grant AUGE/11/14), the Desmoid Tumor Research Foundation, the Desmoid Tumor Foundation of Canada and SOS Desmoïde. We would like to acknowledge Amanda Gonçalves and Benjamin Pavie (VIB Bioimaging Core) for generating the QuPath script for positive cell detection. Furthermore, we are thankful to Tim Deceuninck for the good animal care. Finally, we would like to thank Joeri Tulkens for critical proofreading of the manuscript.
Author information
Authors and Affiliations
Contributions
DT, TP, SG, PVV, and KV designed the study. DT and MB performed all genome engineering experiments. DT, SDem, TN, and MB performed general phenotyping experiments of the leukemia models. DT, MB, and SDew performed RNA expression analysis. WT executed the irradiation procedure. DT and GVI performed flow cytometry experiments. DT, MB, and SDem performed all stainings. TN performed binominal statistics. MC did the BATCH-GE analysis of amplicon deep sequencing data. DC provided histopathological feedback on blinded organ slides. DT and KV wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Tulkens, D., Boelens, M., Naert, T. et al. Mutations in the histone methyltransferase Ezh2 drive context-dependent leukemia in Xenopus tropicalis. Leukemia 37, 2404–2413 (2023). https://doi.org/10.1038/s41375-023-02052-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41375-023-02052-2