Abstract
Isocitrate dehydrogenase 2 (IDH2) mutations occur in more than 15% of cytogenetically normal acute myeloid leukemia (CN-AML) but comparative studies of their roles in leukemogenesis have been scarce. We generated zebrafish models of IDH2R172K and IDH2R140Q AML and reported their pathologic, functional and transcriptomic features and therapeutic responses to target therapies. Transgenic embryos co-expressing FLT3ITD and IDH2 mutations showed accentuation of myelopoiesis. As these embryos were raised to adulthood, full-blown leukemia ensued with multi-lineage dysplasia, increase in myeloblasts and marrow cellularity and splenomegaly. The leukemia cells were transplantable into primary and secondary recipients and resulted in more aggressive disease. Tg(Runx1:FLT3ITDIDH2R172K) but not Tg(Runx1:FLT3ITDIDH2R140Q) zebrafish showed an increase in T-cell development at embryonic and adult stages. Single-cell transcriptomic analysis revealed increased myeloid skewing, differentiation blockade and enrichment of leukemia-associated gene signatures in both zebrafish models. Tg(Runx1:FLT3ITDIDH2R172K) but not Tg(Runx1:FLT3ITDIDH2R140Q) zebrafish showed an increase in interferon signals at the adult stage. Leukemic phenotypes in both zebrafish could be ameliorated by quizartinib and enasidenib. In conclusion, the zebrafish models of IDH2 mutated AML recapitulated the morphologic, clinical, functional and transcriptomic characteristics of human diseases, and provided the prototype for developing zebrafish leukemia models of other genotypes that would become a platform for high throughput drug screening.
Similar content being viewed by others
Introduction
Isocitrate dehydrogenases (IDH) are a group of enzymes that catalyze the conversion of isocitrate to α-ketoglutarate in the physiologic citrate acid cycle [1]. Mutations of IDH include IDH1R132, IDH2R140, and IDH2R172 in which arginine is substituted and they occur in more than 25% cases of acute myeloid leukemia (AML) with normal cytogenetics [2]. IDH mutations confer novel substrate specificity to the enzyme and instead of converting isocitrate to α-ketoglutarate, mutated IDH2 convert the latter to 2-hydroxyglutarate (2-HG). 2-HG is an oncometabolite and is associated with epigenetic alteration, genetic instability, and malignant transformation of hematopoietic cells [3,4,5]. Transgenic and knock-in mouse models of IDH1R132 and IDH2R140Q mutated AML have been reported, showing that these mutations, singly or in combination with co-existing mutations, induced leukemogenesis [6,7,8,9]. However, animal models of mutated IDH2R172K with clinicopathologic characteristics of human AML are scarce [6, 7, 10, 11].
Zebrafish has emerged as a model organism to study human diseases, including leukemia [12]. The optical transparency and high fecundity are distinct advantages, and the zebrafish genome and hematopoietic system are remarkably similar to those in mice and human [13]. Moreover, recent advances in genome editing, transgenesis, and rapid embryonic development have made zebrafish a unique model for studying mutation combinations at high throughput [14]. Over-expression of human IDH1R132H has been shown to induce myelopoiesis in zebrafish embryos, suggesting that the pathogenetic pathway in IDH mutation is conserved in zebrafish [15, 16].
In this study, we established transgenic zebrafish models of IDH2R172K that recapitulated clinicopathologic features of human AML. Comparative studies on IDH2R140Q highlighted both similarities and differences between the two IDH2 mutants in leukemogenesis. These models may provide important platforms for high throughput drug screening targeting IDH mutations in AML.
Results
Effects of human IDH2 mutations on myelopoiesis
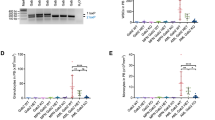
Zebrafish idh2 exhibited remarkable similarities in amino acid sequence and syntenic neighboring genes to those of humans (Fig. S2A, B), suggesting orthologous relationships. To examine the effects of human IDH2 mutations on myelopoiesis at the embryonic stage, IDH2R172K and IDH2R140Q mRNA were microinjected up to 200 pg into wildtype zebrafish embryos at 1-cell stage. Expression of IDH2 mutations induced a marked increase in 2HG level, surrogate of mutant IDH2 expression (Fig. S3A). Intriguingly, the equivalent amount of mRNA induced a substantially higher increase in 2HG in IDH2R172K than in IDH2R140Q injected embryos. Transient expression of IDH2R172K or IDH2R140Q mRNA had little effect on primitive myeloid progenitor as shown by whole mount in-situ hybridization (WISH) of pu.1 (Fig. S3B, C); however, both IDH2 mutations induced a remarkable increase in definitive hematopoietic stem and progenitor cells (HSPC) (cmyb) (Fig. S3B, D) and neutrophils (myeloperoxidase, mpo and Sudan Black B, SBB staining) (Fig. S3B, E, F). The prominent changes in embryonic myelopoiesis induced by IDH2 mutations prompted us to generate transgenic zebrafish lines with stable expression of IDH2 mutations in a lineage-specific manner. The generation of stable transgenic lines was described in “Materials and Methods” and Supplementary Information. F1 transgenic embryos were examined for hematopoietic gene expression by WISH and genotyped individually afterward. Interestingly, lineage-specific expression of IDH2 mutations did not alter the abundance of HSPC or myeloid cells (Fig. 1A–C) in the embryos. On the other hand, only Tg(Runx1:IDH2R172K) but not Tg(Runx1:IDH2R140Q) embryos showed a significant increase in rag1 (T-cell marker) expression compared with wildtype siblings (Fig. 1A, D). F2 double transgenic embryos were generated by crossing F1 Tg(Runx1:IDH2R172K) or Tg(Runx1:IDH2R140Q) to Tg(Runx1:FLT3ITD) [17] to investigate potential synergistic effects of these mutant genes which may co-exist in AML patients. Embryos co-expressing FLT3ITD and IDH2R140Q or IDH2R172K showed a significant increase in markers associated with HSPC (cmyb, runx1) (Fig. 1A, B; Fig. S4A, B); neutrophils (mpo; SBB) (Fig. 1A, C; Fig. S4A, C) and pan-leukocyte marker l-plastin (Fig. S4A, D), when compared with siblings carrying single or no mutation. An increase in T-cell marker (rag1) (Fig. 1A, D) in the developing thymus was only seen in embryos carrying IDH2R172K but not IDH2R140Q irrespective of co-existing FLT3-ITD. Markers associated with primitive erythropoiesis (gata1, hbae1.1) and early myeloid progenitor (pu.1) were unaffected (Fig. S4A, E, F, G).
A Brightfield images of WISH for cmyb at 36hpf, mpo at 48hpf, and rag1 at 5dpf in the mutant (n = 20) and WT (n = 20) zebrafish embryos. B Quantification of cmyb, mpo (C), and rag1 (D) respectively. The expression of each marker was presented as the percentage of each marker expressed in the mutant embryos relative to that of the WT embryos. Data are mean ± s.e.m and statistical analysis was performed by One-way Anova, *P < 0.05, **P < 0.01, ****P < 0.0001.
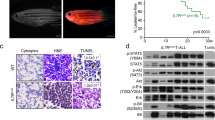
F2 larvae carrying single IDH2 mutation or double mutations with FLT3ITD were raised to adulthood. Morphologically, Tg(Runx1:FLT3ITDIDH2R140Q) and Tg(Runx1:FLT3ITDIDH2R172K) showed circulating blasts in peripheral blood (Fig. 2A, E). Kidney marrow (KM) showed dyserythropoietic features, characterized by the presence of bi-nucleated erythroid cells, nuclear-cytoplasmic asynchrony, and nuclear irregularity (Fig. 2B and Fig. S5). Neutrophils were dysplastic and morphologic features of pseudo-Pelger-Huet anomaly and hypogranularity, reminiscent of human myelodysplastic syndrome, were evident (Fig. 2B and Fig. S5). KM blasts showed abnormal morphology with highly irregular nuclear contours (Fig. 2B and Fig. S5).
A Representative Wright’s staining of the peripheral blood (PB) and KM cells (B), and myeloperoxidase staining of the KM cells (C) from the mutant zebrafish and WT siblings. The blast and Mpo+ cells are indicated by red and black arrows respectively. D Visualization of the thymus from the mutant and WT zebrafish via fluorescent microscopy, the edge of the fish head, thymus and eye was indicated by white dash lines in Tg(rag2:EGFP) background fish. E The percentage of the blast cell in the PB of the transgenic mutant zebrafish (n = 10) and WT siblings (n = 10). F The total cell number, the percentage of blast cell (G), the percentage of Mpo+ cells (H) in the KM of the transgenic mutant zebrafish (n = 6) compared with WT siblings (n = 6). I The relative size of the thymus in the transgenic mutant zebrafish (n = 9) compared with WT siblings (n = 9). J Relative mRNA expression of IDH2, cmyb, rag1, cd9a, pax5, and cd79a in the thymus of the IDH2R172K (n = 3) and wildtype zebrafish (n = 3). K Overall survival of the WT (n = 33), single mutant (n = 33 for FLT3ITD, n = 21 for IDH2R140Q, n = 19 for IDH2R172K), and double mutant (n = 19 for FLT3ITDIDH2R140Q, n = 20 for FLT3ITDIDH2R172K) zebrafish. The numbers in the bottom right-hand corner in A–C indicated the number of fish with the characteristic phenotypes / the total number of fish in each group. Data are mean ± s.e.m. One-way Anova was performed for E, F, G, H, and I, *P < 0.05, **P < 0.01, ****P < 0.0001. Student’s t test was performed for J, **P < 0.01, ***P < 0.001, ****P < 0.0001. Log-Rank test was performed for K, *P < 0.05.
Quantitatively, there were significant increases in total cell number per KM (Fig. 2F) and KM blast percentage [Tg(Runx1:FLT3ITDIDH2R140Q): Blasts = 35.6 ± 2.7%; Tg(Runx1:FLT3ITDIDH2R172K): Blasts = 28.9 ± 4.2%] (Fig. 2B, G), which were positive for Mpo, consistent with their myeloid lineage (Fig. 2C, H). To further quantify KM changes in these transgenic fish, they were crossed to Tg (mpo: EGFP), and EGFP signals were used to define the KM size (Fig. S6A). Both double transgenic mutant fish showed a significant increase in KM size (Fig. S6A, C). Furthermore, there was a significant increase in the size and cellularity of the spleen (Fig. S6B, D–F,), which was infiltrated by blasts and myelomonocytic cells (Fig. S6G).
The increase in rag1 expression in F2 embryos carrying Tg(Runx1:IDH2R172K) and Tg(Runx1:FLT3ITDIDH2R172K) led us to examine T-cell development in adult fish. These embryos were raised to adulthood and crossed with Tg(rag2: EGFP) fish in which T-cells in the thymus expressed EGFP (Fig. 2D). The thymus of the adult transgenic Tg(rag2:EGFP/Runx1:IDH2R172K) and Tg(rag2:EGFP/Runx1:FLT3ITDIDH2R172K) fish were larger than the IDH2R140Q counterparts (Fig. 2D, I). The enlarged thymus expressed mutated human IDH2R172K (Fig. S6H) and showed an increase in rag1 and cmyb (T-cell progenitors and common lymphoid progenitors) but a decrease in cd79a, pax5, and cd9a (B-cells) expression (Fig. 2J).
Double transgenic adult fish also showed a shortened survival after 100 days of age [Median survival: WT: not reach; single mutant: not reach; Tg(Runx1:FLT3ITDIDH2R172K): 221.5 days; Tg(Runx1:FLT3ITDIDH2R140Q): 240 days] (Fig. 2K). The surviving fish also had significant weight loss as they became more mature (Fig. S6I), as compared with their siblings with single or no mutation.
Transplantability of IDH2 R140Q and IDH2 R172K AML cells
To determine the oncogenic potential of IDH2R140Q and IDH2R172K AML in zebrafish, their abilities to propagate upon transplantation were examined using transgenic fish of Tg(Runx1:FLT3ITD) background (Fig. 3A). Lethally irradiated WT adult fish could be rescued by WT KM (Fig. 3B). Tg(Runx1:FLT3ITDIDH2R140Q) and Tg(Runx1:FLT3ITDIDH2R172K) recipients showed a significantly shorter survival [Median survival: WT: not reach; Tg(Runx1:FLT3ITDIDH2R172K): not reach; Tg(Runx1:FLT3ITDIDH2R140Q): 25 days; Not transplanted: 13.5 days] (Fig. 3B). Successful engraftment by leukemic clones was shown by the increase in blasts in the blood (Fig. 3C, D) and KM (Fig. 3E, G) and an increase in cellularity (Fig. 3F) of the latter 30 days post-transplantation. The recipient marrow showed expression of human FLT3 and IDH2, which were not present in the recipients of wildtype marrow (Fig. 3H). Spleen size (Fig. 3I) in the recipients has increased and there were also increases in macrophages and blasts in the spleen of recipients transplanted with Tg(Runx1:FLT3ITDIDH2R172K) and Tg(Runx1: FLT3ITDIDH2R140Q) KM (Fig. 3J, K). Secondary transplantation was also performed. Leukemic engraftment was significantly more aggressive [Median survival: WT: not reach; Tg(Runx1:FLT3ITDIDH2R172K): 15 days; Tg(Runx1:FLT3ITDIDH2R140Q): 13.5 days] (Fig. 3L).
A Schematic representation of the transplantation procedure. B Survival plots of the irradiated recipients without transplantation (n = 6) or transplanted with KM cells from double transgenic mutant zebrafish (n = 32 for FLT3ITDIDH2R140Q, n = 25 for FLT3ITDIDH2R172K) or WT(n = 21) 40 days post-primary transplantation. C The percentage of the blast cell in the PB and the representative Wright’s staining of the PB (D) of the primary recipients (n = 6 for all groups) 30 days post-transplantation. E The percentage of the blast cell and the total cell number (F) in the KM and the representative Wright’s staining of the KM (G) of the primary recipients (n = 6 for all groups) 30 days post-transplantation. H Relative mRNA expression of FLT3 and IDH2 in the KM of the primary recipients (n = 3 for all groups). I The size and the representative Wright’s staining of the spleen (J), and the percentage of the blast cell and macrophages in the spleens (K) of the primary recipients (n = 5 for all groups) 30 days post-transplantation. Blast cells and macrophages are indicated by red and black arrows respectively. L Survival plot of the irradiated secondary recipients (n = 16 for FLT3ITDIDH2R140Q, n = 15 for others) 40 days post-transplantation. Data are mean ± s.e.m. Student’s t test was performed for C, E, F, H, I, and K (each mutant group vs. WT), **P < 0.01, ***P < 0.001, ****P < 0.0001. Log-Rank test was performed for B and L, *P < 0.05, **P < 0.01, ****P < 0.0001.
Co-expression of FLT3 ITD and IDH2 mutation promoted leukemogenesis at the transcriptional level
To investigate the cellular compartments in Tg(Runx1:FLT3ITDIDH2R172K) and Tg(Runx1:FLT3ITDIDH2R140Q) transgenic zebrafish, single-cell RNA-sequencing was performed (Fig. 4A). After removing doublets and filtering by sequencing quality control, 34,214 cells were included in subsequent analyses. Transcriptomes from WT and transgenic mutant KM cells were pooled and clustered using R Seurat Package, resulting in 16 major cell populations (Fig. 4B, C), which were identified by previously reported lineage-specific marker genes [18,19,20,21,22,23,24,25,26,27,28] (Fig. S7A) and were visualized using the Uniform Manifold Approximation and Projection (UMAP) approach (https://arxiv.org/abs/1802.03426) (Fig. 4B). KM from both transgenic fish showed an increase in the prevalence of CLP, macrophage, HSPC-MPP, CMP, thrombocyte, erythroid progenitor, B-cell and HSC populations but a decrease in erythrocyte population when compared with WT siblings (Fig. 4D), indicating multi-lineage hematopoietic expansion by combined expression of IDH2 mutations and FLT3ITD when they were expressed in HSPC as driven by the Runx1 enhancer and mouse β-globin minimal promoter.
A Schematic representation of the single-cell RNA sequencing experimental workflow. B 16 distinct cell clusters were visualized by Uniform manifold approximation projection (UMAP). C Cell type classification based on the key hematopoietic markers identified in zebrafish. D Quantitation of proportions of distribution between double mutant and WT KM cells in different clusters. E Sub-clustering of HSPC-MPP population. F Quantitation of proportions of distribution between double mutant and WT zebrafish in the different sub-clusters of HSPC-MPP. The red dot represents the difference of the Log2FC of the proportion of a specific cluster in Tg(Runx1:FLT3ITDIDH2R140Q) compared to that of WT is over 0.25 or less than −0.25, the blue dot represents the difference of the Log2FC of the proportion of a specific cluster in Tg(Runx1:FLT3ITDIDH2R172K) compared to that of WT is over 0.25 or less than −0.25, and the gray dot indicates no change (−0.25 < log2FC < 0.25) was found in the proportion of a specific cluster when compared between the double mutant and WT zebrafish.
To investigate the mechanisms of myeloid leukemogenesis in the transgenic fish, the HSPC-MPP populations were further examined for possible lineage skewing. Eight sub-clusters were identified, including three myeloid subclusters with up-regulation of myeloid gene s100a10b, one HSC subcluster characterized by myb expression and four lymphoid subclusters with increased expression of igic1s1 (Fig. 4E and Fig. S7B). Both mutant IDH2 transgenic fish showed a significant increase in the prevalence of myeloid subclusters and a significant decrease of lymphoid subclusters (Fig. 4F), suggestive of HSPC-MPP priming towards a myeloid fate.
To examine the effects of IDH2 mutations on lineage differentiation, lineage trajectory and pseudotime analyses were performed for erythroid and myeloid lineages (Fig. 5A–H). Lineage-specific progenitors and terminally differentiated mature cells were differentiated from HSPC-MPP, with increasing pseudotime values along differentiation (Fig. 5A, B, E and F). Pseudotime of lineage-specific progenitors and erythrocytes from Tg(Runx1:FLT3ITDIDH2R140Q) and Tg(Runx1:FLT3ITDIDH2R172K), and that of neutrophils from Tg(Runx1:FLT3ITDIDH2R140Q), were significantly lower than those of their WT siblings (Fig. 5C, D, G and H). Interestingly, while these mutation combinations induced myeloid priming of HSPC-MPP, as shown by a significant increase in the prevalence of myeloid subclusters and significant decrease of lymphoid subclusters within this population, they also induced differentiation blockage downstream of HSPC-MPP, as shown by the lower pseudotime in the downstream populations.
A Lineage differentiation trajectory and pseudotime (B) analysis for the erythroid lineage, with HSPC-MPP as the starting point. C Quantification of the pseudotime values of the erythroid progenitor and erythrocyte (D) in the double mutant and wildtype zebrafish. E Lineage differentiation trajectory and pseudotime (F) analysis for the myeloid lineage, with HSPC-MPP as the starting point. G Quantification of the pseudotime values of the myeloid progenitor and neutrophil (H) in the double mutant and WT zebrafish. I Differentially enriched Hallmark and C2 pathway signatures within the HSC, HSPC-MPP, myeloid, and erythroid progenitor clusters between Tg(Runx1:FLT3ITDIDH2R140Q) and WT or Tg(Runx1:FLT3ITDIDH2R172K) and WT. Wilcoxon Rank Sum test was performed for C, D, G, and H, ****P < 0.0001.
To further evaluate the effects of IDH2 mutations on the initiation and promotion of leukemogenesis, gene set enrichment analysis (GSEA) was performed for early hematopoietic cell populations, including HSC, HSPC, myeloid, and erythroid progenitor cell clusters, based on differentially expressed genes between Tg(Runx1:FLT3ITDIDH2R172K), Tg(Runx1:FLT3ITDIDH2R140Q) and their WT siblings (Fig. 5I). In both double transgenic fish, genes associated with MTORC, MYC and RAS signaling, were positively enriched in most cell clusters, consistent with their leukemia phenotypes. Intriguingly, the Tg(Runx1:FLT3ITDIDH2R172K) zebrafish showed positive enrichment of genes associated with interferon responses and signaling whereas the Tg(Runx1:FLT3ITDIDH2R140Q) zebrafish showed positive enrichment of genes associated with interleukin 1-related signaling.
Use of transgenic fish in therapeutic evaluation
The clinical relevance of the zebrafish models was tested at both embryonic and adult stages. Tg(Runx1:FLT3ITDIDH2R172K) and Tg(Runx1:FLT3ITDIDH2R140Q) embryos and their WT siblings were treated with gilteritinib and quizartinib (FLT3 inhibitors) as well as enasidenib (IDH2 inhibitor), which have been shown to induce clinical response and confer a survival advantage to patients with FLT3ITD and IDH2 mutations (Fig. 6A). Using cytochemical staining with SBB as a surrogate for embryonic myelopoiesis, these inhibitors ameliorated the increase in myelopoiesis in the double transgenic embryos (Fig. 6B–E). The therapeutic responses were also tested in adult fish (Fig. 6F). Initial dose-finding studies showed that daily gavage of quizartinib at 10 mg/kg and enasidenib at 100 mg/kg were compatible with normal fish survival (Fig. S8A, B). Double transgenic leukemic fish and their WT siblings were treated with 14 days of quizartinib, enasidenib, or their combination, and their KM cellularity, blast, neutrophil, and erythrocyte counts were enumerated (Fig. 6G–K). Quizartinib but not enasidenib monotherapy significantly reduced cellularity in the KM of the double transgenic fish (Fig. 6G). Enasidenib but not quizartinb monotherapy significantly increased the percentage of neutrophil and erythrocyte and slightly decreased the blast cell population in the KM of the double transgenic fish (Fig. S8C–I). Combination treatment significantly reduced blast population (Fig. 6H, I), increased neutrophil abundance (Fig. 6H, J), and restored erythropoiesis in the KM (Fig. 6H, K). When the effects of these therapeutic agents on the spleen were examined, only the combination of quizartinib and enasidenib effectively reduced the size of the spleen (Fig S8J, K).
A Schematic representation of drug administration and assessment in zebrafish embryos. B Representative SBB staining in the double mutant and WT embryos following in vivo treatment of vehicle control, gilteritinib, quizartinib, and enasidenib at 0.01 μM, 0.1 μM and 1 μM respectively. C–E Quantification of SBB + cells, presented as the percentage of SBB + cells of treated zebrafish for relative to untreated WT control (n = 20 for all groups) in B. F Schematic representation of drug administration and assessment in adult zebrafish. G Changes in total KM cellularity in the adult double mutant and WT zebrafish post-treatment. H Representative Wright’s staining of the KM from the adult double mutant and WT zebrafish 14 days post-treatment. I The percentage of blast cell, neutrophil (J), and erythrocyte (K) in the KM of double mutant and WT zebrafish (n = 5 for all groups). Data are mean ± s.e.m and statistical analysis was performed by Student’s t test (treated vs. untreated for each genotype), *P < 0.05, **P < 0.01, ***P < 0.001.
Discussion
IDH2R172K and IDH2R140Q mutations occur in more than 15% of patients with cytogenetically normal AML. Despite their frequent occurrence, comparative studies of their roles in leukemogenesis have been scarce. In this study, we generated transgenic zebrafish models and demonstrated that transgenic expression of IDH2R172K and IDH2R140Q in hematopoietic stem/progenitor cells induced myeloid skewing and differentiation blockade at HSPC-MPP levels, resulting in expansion of KM, splenomegaly, myelodysplasia and increase in Mpo+ blasts capable of self-renewal in serial transplantations. The leukemic phenotypes responded to target therapies expected of their mechanisms of action, attesting to the clinical relevance of the zebrafish models and their potential application in the development of personalized medicine. These findings were consistent with mouse models of transgenic or knockin IDH2R140Q [7, 9, 29, 30], where IDH2 mutation was shown to induce differentiation block and leukemogenesis, either singly or in combination with other genetic perturbations. Moreover, observations arising from this study may shed important lights to our understanding of leukemogenesis pertinent to IDH2 mutation.
First, our observations of IDH2R172K AML in zebrafish were consistent with those in viral transduction studies where IDH2R172K expression in hematopoietic cells induced leukemogenesis [6, 7] but was different from the knock-in mouse model in which IDH2R172K expression in hematopoietic cells led to perturbed lymphoid development but not leukemia [10, 11]. Different experimental models and tissue promoters chosen for IDH2R172K expression as well as cooperative mutation partners might account for the difference in phenotypes. In zebrafish, IDH2R172K alone was found to induce leukemia-like phenotypes in a small percentage of the transgenic zebrafish, and full-blown leukemia was developed in those fish with IDH2R172K in the combination of FLT3ITD. Their cooperativity was also shown by the superior pharmacologic responses to the combination of enasidenib and quizartinib, inhibitors of IDH2R172K and FLT3ITD, underscoring the pathogenetic role of each mutant gene in this model.
Second, we demonstrated that in addition to IDH2R140Q, transgenic expression of IDH2R172K in HSPC induced differentiation blockade of hematopoiesis, as illustrated by trajectory analyses of the single-cell transcriptome. The lower pseudotime of lineage-specific progenitors and mature neutrophils and erythrocytes as they differentiated from HSPC-MPP supported the proposition of differentiation blockade in both IDH2 mutant AML. Furthermore, the rapid recovery of neutrophils and erythrocytes upon treatment with enasidenib and its combination with quizartinib also suggested a relief of differentiation blockade reminiscent of patient response to enasidenib.
Third, single-cell transcriptome analysis of the KM has empowered us to examine the lineage development and hematopoietic phenotypes of zebrafish AML in detail. Both Tg(Runx1:FLT3ITDIDH2R172K) and Tg(Runx1:FLT3ITDIDH2R140Q) showed multi-lineage hematopoietic expansion in KM as compared with wildtype siblings. Expression of these transgenes in HSPC as driven by the Runx1 enhancer and mouse β-globin minimal promoter could lead to clonal expansion of both lymphoid and myeloid progenitors, notably CLP and CMP. A close examination of HSPC-MPP showed full-blown lineage skewing typical of myeloid neoplasm, suggesting leukemogenesis might begin at the HSPC-MPP stage in the zebrafish model. Moreover, GSEA analysis revealed potential associations between Tg(Runx1:FLT3ITDIDH2R172K), Tg(Runx1:FLT3ITDIDH2R140Q) and immune activation with particular reference to interferon and IL-1 signal activation. Their mechanistic links would have to be further evaluated.
Finally, information arising from this study has shed important lights to the hitherto undescribed hematopoietic effects of IDH2 mutations. In particular, IDH2R172K but not IDH2R140Q, accentuated T-cell development at both embryonic and adult stages. The effects were cell-autonomous and IDH2R172K mutation could be demonstrated in the thymus. It was unclear if the preferential effects of IDH2R172K on T-cell development were related to a significantly higher level of oncometabolite 2HG, as demonstrated in zebrafish embryos and mice [10]. To our knowledge, IDH2R172K has not been reported in precursor T-cell lymphoblastic leukemia. Whether the IDH2R172K transgenic zebrafish would provide a disease model of mature T-cell neoplasm, including angioimmunoblastic T-cell lymphoma, in which IDH2R172K mutation occurs in 20% of cases [31,32,33], would have to be further investigated [10].
The zebrafish model was of clinical relevance. As proof-of-principle, both zebrafish embryos and adults carrying FLT3ITDIDH2R172K and FLT3ITDIDH2R140Q mutations responded to quizartinib/gilteritinib, and enasidenib, which were effective agents for FLT3ITD [34, 35] and IDH2 mutant AML [36, 37]. The optical transparency, high fecundity and relatively simple husbandry of zebrafish have made it uniquely suitable for high throughput drug screening. Furthermore, the availability of lineage-specific fluorescent reporter lines has also facilitated the evaluation of drug effects on specific lineages. Intriguingly, increased HSPC and enhanced primitive myelopoiesis were observed only in monogenic IDH2R172K and IDH2R140Q expression in the transient but not stable transgenic model. The apparent discrepancy might be explained by a ubiquitous and higher level of mutant gene expression in the transient system. Whether the transient expression system might provide a more robust high throughput model for zebrafish drug screening would have to be further tested.
In conclusion, the present study generated zebrafish models of AML carrying FLT3ITDIDH2R172K and FLT3ITDIDH2R140Q mutations, recapitulating the morphologic, clinical, transcriptomic and functional characteristics of the corresponding human diseases. These double transgenic fish will become prototypes of zebrafish AML models carrying mutation combinations and powerful tools for rapid drug discovery targeting specific drive mutations.
Methods
Generation of transgenic zebrafish lines
Tol2 transgenesis was used to generate single transgenic fish lines Tg(Runx1: IDH2R140Q) and Tg(Runx1: IDH2R172K) in which human IDH2R140Q or IDH2R172K were expressed under the control of the HSPC-specific Runx1 + 23 enhancer and mouse β-globin minimal promoter (Fig. S1A) [38]. Human sequence of IDH2R140Q or IDH2R172K was cloned into pDONR221 vector to generate the “middle” clone (pME-IDH2 R140Q or IDH2 R172K) by Gateway BP reaction. The final Tol2 integrable construct was generated via multisite Gateway LR reaction in which three entry clones [p5E‐Runx1 + 23 (Addgene #69602), pME‐ IDH2R140Q or IDH2R172K, and p3E‐mCherrypA] [39] were incorporated into the destination vector pDestTol2CG [39] with EGFP fluorescent protein expressed under the control of the cardiomyocyte-specific cmlc2 promoter. The latter served as a marker of successful transgenesis. Single transgenic lines were generated by co-injecting 50–100 pg of the respective Tol2 construct and the Tol2 transposase mRNA into the wildtype (WT) TU embryos at the one-cell stage. Founders were identified by PCR and EGFP fluorescence of the heart. F1 single transgenic fish were generated by outcrossing the identified founder fish with WT TU fish. F1 embryos with positive heart EGFP fluorescence at 2dpf were raised to adulthood (Fig. S1B), and the genotype was further confirmed by genotyping (Fig. S1C). The generation of Tg(Runx1:FLT3ITD) transgenic line was described previously [17]. Double transgenic fish Tg(Runx1:FLT3ITDIDH2R140Q) and Tg(Runx1:FLT3ITDIDH2R172K) were generated by crossing the single transgenic lines of Tg(Runx1: IDH2R140Q/R172K) with Tg(Runx1:FLT3ITD). The expression of FLT3 and IDH2 mRNA in F1 adult fish was confirmed by q-PCR(Fig. S1D). F1 transgenic fish were also outcrossed with different transgenic reporter lines, including Tg(rag2:EGFP) and Tg(mpo: EGFP) for evaluation of thymus and kidney marrow size.
Zebrafish kidney marrow transplantation
WT recipient fish were irradiated at 25 Gy 2 days prior to transplantation. On the day of transplantation, donor KM cells were collected from Tg(Runx1: FLT3ITDIDH2R172K), Tg(Runx1: FLT3ITDIDH2R140Q) or WT fish in 0.9X PBS with 5% FBS buffer, resuspended in injection medium (0.9X PBS, 5% FBS, 0.5 U/μL heparin and 0.2 U/μL Dnase I) to a final concentration of 2 × 105 cells/μL [40] Recipient fish was anesthetized using 0.015% Tricaine and 5 × 105 donor cells were transplanted via intracardiac injection using capillary glass needle (GC100TF-15, Warner Instruments, USA). After injection, recipients were immediately returned to sterilized fish water. Survival of recipients was monitored and recorded daily.
Single-cell RNA sequencing
Sample preparation, library preparation, sequencing, and data analysis details are provided in the Supplementary materials and methods. A total of 34,214 KM cells passed the quality control and were analyzed using Seurat (v4.0.6) [41]. FindClusters were used to determine cell clusters. Marker genes for each cluster were inspected and cluster identities were determined based on previously reported lineage-specific marker genes in zebrafish [18,19,20,21,22,23,24,25,26,27,28]. Differentially expressed genes were analyzed based on their known functions according to the curated hallmark and C2 gene sets from Molecular Signatures Database (MSigDB v7.4 [42, 43] https://www.gsea-msigdb.org/gsea/msigdb/index.jsp). Lineage trajectory and pseudotime analysis were performed by Monocle 2 [44, 45].
Drug treatment on embryos and adult fish
Tg(Runx1:FLT3ITDIDH2R172K), Tg(Runx1:FLT3ITDIDH2R140Q) and WT zebrafish embryos were treated with FLT3 inhibitors Gilteritinib (Selleck S7754), Quizartinib (MedChemExpress HY-13001), or IDH2 inhibitor Enasidenib (MedChemExpress HY-18690) at three concentrations (0.01, 0.1, and 1 μM) in E3 medium with 1-phenyl-2-thiourea at 6 hpf. The medium was changed every 24 h. At 4 dpf, embryos were collected and fixed in 4%PFA for SBB staining. Adult zebrafish were anesthetized using 0.015% Tricaine, propped vertically in a damp sponge, and 5–10 μl of quizartinib (10 mg/kg), enasidenib (100 mg/kg) or their combination, dissolved in H2O containing 10% DMSO (Sigma) and 0.01% Phenol Red (Sigma-Aldrich), were given via oral gavage [46, 47]. Treated zebrafish were placed into individual recovery tank with fresh and sterile fish water immediately. Treatment was given daily for 14 days.
Statistical analysis
Data were assessed for normal distribution with a Shapiro–Wilk normality test using Prism9 (GraphPad, San Diego, CA) and presented as mean ± standard error of the mean (s.e.m) of at least three independent experiments. Sample sizes were determined by power analysis to provide sufficient statistical power to detect differences. Comparisons between group of data were evaluated by two-sided Student’s t test, one-way ANOVA, or Wilcoxon Rank Sum test if the data were normally distributed and the variance was equal. Survival data were evaluated by Kaplan-Meier analyses and compared using Log-Rank test. P value less than 0.05 was considered statistically significant.
Additional methods and materials used in this study are provided in the Supplementary Information.
Data availability
Single-cell RNA sequencing data have been deposited in the National Center for Biotechnology Information Sequence Read Archive under accession number PRJNA911847.
Change history
15 March 2023
A Correction to this paper has been published: https://doi.org/10.1038/s41388-023-02664-z
References
Stoddard BL, Dean A, Koshland DE Jr. Structure of isocitrate dehydrogenase with isocitrate, nicotinamide adenine dinucleotide phosphate, and calcium at 2.5-A resolution: a pseudo-Michaelis ternary complex. Biochemistry. 1993;32:9310–6.
Tsui SP, Ip HW, Saw NY, Zhang C, Cheung AK, Ng NK, et al. Redefining prognostication of de novo cytogenetically normal acute myeloid leukemia in young adults. Blood Cancer J. 2020;10:104.
Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–44.
Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, Coller HA, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17:225–34.
Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19:17–30.
Chen C, Liu Y, Lu C, Cross JR, Morris JP, Shroff AS, et al. Cancer-associated IDH2 mutants drive an acute myeloid leukemia that is susceptible to Brd4 inhibition. Genes Dev. 2013;27:1974–85.
Yoshimi A, Lin KT, Wiseman DH, Rahman MA, Pastore A, Wang B, et al. Coordinated alterations in RNA splicing and epigenetic regulation drive leukaemogenesis. Nature. 2019;574:273–7.
McKenney AS, Lau AN, Somasundara AVH, Spitzer B, Intlekofer AM, Ahn J, et al. JAK2/IDH-mutant-driven myeloproliferative neoplasm is sensitive to combined targeted inhibition. J Clin Investig. 2018;128:789–804.
Kats LM, Reschke M, Taulli R, Pozdnyakova O, Burgess K, Bhargava P, et al. Proto-oncogenic role of mutant IDH2 in leukemia initiation and maintenance. Cell Stem Cell. 2014;14:329–41.
Lemonnier F, Cairns RA, Inoue S, Li WY, Dupuy A, Broutin S, et al. The IDH2 R172K mutation associated with angioimmunoblastic T-cell lymphoma produces 2HG in T cells and impacts lymphoid development. Proc Natl Acad Sci USA. 2016;113:15084–9.
Chen TC, Yao CY, Chen YR, Yuan CT, Lin CC, Hsu YC, et al. Oncogenesis induced by combined Phf6 and Idh2 mutations through increased oncometabolites and impaired DNA repair. Oncogene. 2022;41:1576–88.
Patton EE, Zon LI, Langenau DM. Zebrafish disease models in drug discovery: from preclinical modelling to clinical trials. Nat Rev Drug Discov. 2021;20:611–28.
Wattrus SJ, Zon LI. Blood in the water: recent uses of zebrafish to study myeloid biology. Curr Opin Hematol. 2021;28:43–9.
Shi X, He BL, Ma AC, Leung AY. Fishing the targets of myeloid malignancies in the era of next generation sequencing. Blood Rev. 2016;30:119–30.
Trompouki E. Fish provide ID(H)eas on targeting leukemia. Blood. 2015;125:2880–2.
Shi X, He BL, Ma AC, Guo Y, Chi Y, Man CH, et al. Functions of idh1 and its mutation in the regulation of developmental hematopoiesis in zebrafish. Blood. 2015;125:2974–84.
He BL, Yang N, Man CH, Ng NK, Cher CY, Leung HC, et al. Follistatin is a novel therapeutic target and biomarker in FLT3/ITD acute myeloid leukemia. EMBO Mol Med. 2020;12:e10895.
Athanasiadis EI, Botthof JG, Andres H, Ferreira L, Lio P, Cvejic A. Single-cell RNA-sequencing uncovers transcriptional states and fate decisions in haematopoiesis. Nat Commun. 2017;8:2045.
Avagyan S, Weber MC, Ma S, Prasad M, Mannherz WP, Yang S, et al. Single-cell ATAC-seq reveals GATA2-dependent priming defect in myeloid and a maturation bottleneck in lymphoid lineages. Blood Adv. 2021;5:2673–86.
Chestnut B, Casie Chetty S, Koenig AL, Sumanas S. Single-cell transcriptomic analysis identifies the conversion of zebrafish Etv2-deficient vascular progenitors into skeletal muscle. Nat Commun. 2020;11:2796.
Gioacchino E, Koyunlar C, Zink J, de Looper H, de Jong M, Dobrzycki T, et al. Essential role for Gata2 in modulating lineage output from hematopoietic stem cells in zebrafish. Blood Adv. 2021;5:2687–700.
Lawson ND, Li R, Shin M, Grosse A, Yukselen O, Stone OA, et al. An improved zebrafish transcriptome annotation for sensitive and comprehensive detection of cell type-specific genes. Elife. 2020;9:e55792.
Macaulay IC, Svensson V, Labalette C, Ferreira L, Hamey F, Voet T, et al. Single-cell RNA-sequencing reveals a continuous spectrum of differentiation in hematopoietic cells. Cell Rep. 2016;14:966–77.
Moore FE, Garcia EG, Lobbardi R, Jain E, Tang Q, Moore JC, et al. Single-cell transcriptional analysis of normal, aberrant, and malignant hematopoiesis in zebrafish. J Exp Med. 2016;213:979–92.
Olsson A, Venkatasubramanian M, Chaudhri VK, Aronow BJ, Salomonis N, Singh H, et al. Single-cell analysis of mixed-lineage states leading to a binary cell fate choice. Nature. 2016;537:698–702.
Xia J, Kang Z, Xue Y, Ding Y, Gao S, Zhang Y, et al. A single-cell resolution developmental atlas of hematopoietic stem and progenitor cell expansion in zebrafish. Proc Natl Acad Sci USA. 2021;118:e2015748118.
Yakulov TA, Todkar AP, Slanchev K, Wiegel J, Bona A, Gross M, et al. CXCL12 and MYC control energy metabolism to support adaptive responses after kidney injury. Nat Commun. 2018;9:3660.
Tang Q, Iyer S, Lobbardi R, Moore JC, Chen H, Lareau C, et al. Dissecting hematopoietic and renal cell heterogeneity in adult zebrafish at single-cell resolution using RNA sequencing. J Exp Med. 2017;214:2875–87.
Zhang X, Wang X, Wang XQD, Su J, Putluri N, Zhou T, et al. Dnmt3a loss and Idh2 neomorphic mutations mutually potentiate malignant hematopoiesis. Blood. 2020;135:845–56.
Shih AH, Meydan C, Shank K, Garrett-Bakelman FE, Ward PS, Intlekofer AM, et al. Combination targeted therapy to disrupt aberrant oncogenic signaling and reverse epigenetic dysfunction in IDH2- and TET2-mutant acute myeloid leukemia. Cancer Discov. 2017;7:494–505.
Cairns RA, Iqbal J, Lemonnier F, Kucuk C, de Leval L, Jais JP, et al. IDH2 mutations are frequent in angioimmunoblastic T-cell lymphoma. Blood. 2012;119:1901–3.
Steinhilber J, Mederake M, Bonzheim I, Serinsoz-Linke E, Muller I, Fallier-Becker P, et al. The pathological features of angioimmunoblastic T-cell lymphomas with IDH2(R172) mutations. Mod Pathol. 2019;32:1123–34.
Wang C, McKeithan TW, Gong Q, Zhang W, Bouska A, Rosenwald A, et al. IDH2R172 mutations define a unique subgroup of patients with angioimmunoblastic T-cell lymphoma. Blood. 2015;126:1741–52.
Cortes JE, Khaled S, Martinelli G, Perl AE, Ganguly S, Russell N, et al. Quizartinib versus salvage chemotherapy in relapsed or refractory FLT3-ITD acute myeloid leukaemia (QuANTUM-R): a multicentre, randomised, controlled, open-label, phase 3 trial. Lancet Oncol. 2019;20:984–97.
Perl AE, Martinelli G, Cortes JE, Neubauer A, Berman E, Paolini S, et al. Gilteritinib or chemotherapy for relapsed or refractory FLT3-mutated AML. N Engl J Med. 2019;381:1728–40.
Stein EM, DiNardo CD, Pollyea DA, Fathi AT, Roboz GJ, Altman JK, et al. Enasidenib in mutant IDH2 relapsed or refractory acute myeloid leukemia. Blood. 2017;130:722–31.
Yen K, Travins J, Wang F, David MD, Artin E, Straley K, et al. AG-221, a first-in-class therapy targeting acute myeloid leukemia harboring oncogenic IDH2 mutations. Cancer Discov. 2017;7:478–93.
Tamplin OJ, Durand EM, Carr LA, Childs SJ, Hagedorn EJ, Li P, et al. Hematopoietic stem cell arrival triggers dynamic remodeling of the perivascular niche. Cell. 2015;160:241–52.
Kwan KM, Fujimoto E, Grabher C, Mangum BD, Hardy ME, Campbell DS, et al. The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev Dyn. 2007;236:3088–99.
Fang X, Xu S, Zhang Y, Xu J, Huang Z, Liu W, et al. Asxl1 C-terminal mutation perturbs neutrophil differentiation in zebrafish. Leukemia. 2021;35:2299–310.
Hao Y, Hao S, Andersen-Nissen E, Mauck WM 3rd, Zheng S, Butler A, et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184:3573–87. e29
Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P. The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–25.
Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinforma. 2011;27:1739–40.
Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014;32:381–6.
Qiu X, Mao Q, Tang Y, Wang L, Chawla R, Pliner HA, et al. Reversed graph embedding resolves complex single-cell trajectories. Nat Methods. 2017;14:979–82.
Collymore C, Rasmussen S, Tolwani RJ. Gavaging adult zebrafish. J Vis Exp. 2013;11:50691.
Dang M, Henderson RE, Garraway LA, Zon LI. Long-term drug administration in the adult zebrafish using oral gavage for cancer preclinical studies. Dis Model Mech. 2016;9:811–20.
Acknowledgements
We thank the Zebrafish Facility from the Centre for Comparative Medicine Research (CCMR) at The University of Hong Kong. We thank Carol Y.K. Yiu, Timothy C. C. Ng, and May Chu for animal assistance. The works were supported by Theme-based Research Scheme (T12-702/20-N) and Health and Medical Research Fund Project No. 03144046 and Project No. 06173456.
Author information
Authors and Affiliations
Contributions
DW, AYHL, and XS conceptualized the study and designed the experiments. DW, BYLC, and SPT performed the experiments. LZ and XY performed the bioinformatics analysis. DW, AYHL, and XS analyzed the data and wrote the manuscript. CFS, RL, ACM, and BLH contributed intellectually. All authors approved and contributed to the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wang, D., Zheng, L., Cheng, B.Y.L. et al. Transgenic IDH2R172K and IDH2R140Q zebrafish models recapitulated features of human acute myeloid leukemia. Oncogene 42, 1272–1281 (2023). https://doi.org/10.1038/s41388-023-02611-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-023-02611-y