Abstract
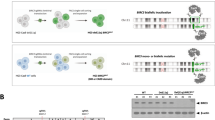
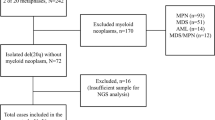
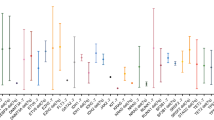
Chronic lymphocytic leukemia (CLL) is a heterogeneous disease, the prognosis of which varies according to the cytogenetic group. We characterized a rare chromosomal abnormality (del(8p), deletion of the short arm of chromosome 8) in the context of CLL. By comparing the largest cohort of del(8p) CLL to date (n = 57) with a non-del(8p) cohort (n = 155), del(8p) was significantly associated with a poor prognosis, a shorter time to first treatment, worse overall survival (OS), and a higher risk of Richter transformation. For patients treated with fludarabine-based regimens, the next-treatment-free survival and the OS were shorter in del(8p) cases (including those with mutated IGHV). One copy of the TNFRSF10B gene (coding a pro-apoptotic receptor activated by TRAIL) was lost in 91% of del(8p) CLL. TNFRSF10B was haploinsufficient in del(8p) CLL, and was involved in the modulation of fludarabine-induced cell death - as confirmed by our experiments in primary cells and in CRISPR-edited TNFRSF10B knock-out CLL cell lines. Lastly, del(8p) abrogated the synergy between fludarabine and TRAIL-induced apoptosis. Our results highlight del(8p)’s value as a prognostic marker and suggest that fit CLL patients (i.e. with mutated IGHV and no TP53 disruption) should be screened for del(8p) before the initiation of fludarabine-based treatment.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Data are available within the manuscript, figures, or supplementary information. The datasets generatedand/or analysed during the current study are available from the corresponding author on reasonable request.
References
Brown JR, Hanna M, Tesar B, Werner L, Pochet N, Asara JM, et al. Integrative genomic analysis implicates gain of PIK3CA at 3q26 and MYC at 8q24 in chronic lymphocytic leukemia. Clin Cancer Res: Off J Am Assoc Cancer Res. 2012;18:3791–802.
Forconi F, Rinaldi A, Kwee I, Sozzi E, Raspadori D, Rancoita PM, et al. Genome-wide DNA analysis identifies recurrent imbalances predicting outcome in chronic lymphocytic leukaemia with 17p deletion. Br J Haematol. 2008;143:532–6.
Bea S, Lopez-Guillermo A, Ribas M, Puig X, Pinyol M, Carrio A, et al. Genetic imbalances in progressed B-cell chronic lymphocytic leukemia and transformed large-cell lymphoma (Richter’s syndrome). Am J Pathol. 2002;161:957–68.
Houldsworth J, Guttapalli A, Thodima V, Yan XJ, Mendiratta G, Zielonka T, et al. Genomic imbalance defines three prognostic groups for risk stratification of patients with chronic lymphocytic leukemia. Leuk Lymphoma. 2014;55:920–8.
Leeksma AC, Baliakas P, Moysiadis T, Puiggros A, Plevova K, Van der Kevie-Kersemaekers AM, et al. Genomic arrays identify high-risk chronic lymphocytic leukemia with genomic complexity: a multi-center study. Haematologica. 2021;106:87–97.
Landau DA, Carter SL, Stojanov P, McKenna A, Stevenson K, Lawrence MS, et al. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell. 2013;152:714–26.
Landau DA, Tausch E, Taylor-Weiner AN, Stewart C, Reiter JG, Bahlo J, et al. Mutations driving CLL and their evolution in progression and relapse. Nature. 2015;526:525–30.
Rinaldi A, Mian M, Kwee I, Rossi D, Deambrogi C, Mensah AA, et al. Genome-wide DNA profiling better defines the prognosis of chronic lymphocytic leukaemia. Br J haematol. 2011;154:590–9.
Blanco G, Puiggros A, Baliakas P, Athanasiadou A, Garcia-Malo M, Collado R, et al. Karyotypic complexity rather than chromosome 8 abnormalities aggravates the outcome of chronic lymphocytic leukemia patients with TP53 aberrations. Oncotarget. 2016;7:80916–24.
Burger JA, Landau DA, Taylor-Weiner A, Bozic I, Zhang H, Sarosiek K, et al. Clonal evolution in patients with chronic lymphocytic leukaemia developing resistance to BTK inhibition. Nat Commun. 2016;7:11589.
Rubio-Moscardo F, Blesa D, Mestre C, Siebert R, Balasas T, Benito A, et al. Characterization of 8p21.3 chromosomal deletions in B-cell lymphoma: TRAIL-R1 and TRAIL-R2 as candidate dosage-dependent tumor suppressor genes. Blood. 2005;106:3214–22.
Hallek M, Cheson BD, Catovsky D, Caligaris-Cappio F, Dighiero G, Dohner H, et al. iwCLL guidelines for diagnosis, indications for treatment, response assessment, and supportive management of CLL. Blood. 2018;131:2745–60.
Baliakas P, Jeromin S, Iskas M, Puiggros A, Plevova K, Nguyen-Khac F, et al. Cytogenetic complexity in chronic lymphocytic leukemia: definitions, associations, and clinical impact. Blood. 2019;133:1205–16.
Cosson A, Chapiro E, Belhouachi N, Cung H-A, Keren B, Damm F, et al. 14q deletions are associated with trisomy 12, NOTCH1 mutations and unmutated IGHV genes in chronic lymphocytic leukemia and small lymphocytic lymphoma. Genes Chromosom Cancer. 2014;53:657–66.
Hertlein E, Beckwith KA, Lozanski G, Chen TL, Towns WH, Johnson AJ, et al. Characterization of a new chronic lymphocytic leukemia cell line for mechanistic in vitro and in vivo studies relevant to disease. PloS ONE. 2013;8:e76607.
Cao J, Wu L, Zhang SM, Lu M, Cheung WK, Cai W, et al. An easy and efficient inducible CRISPR/Cas9 platform with improved specificity for multiple gene targeting. Nucleic Acids Res. 2016;44:e149.
Boudny M, Zemanova J, Khirsariya P, Borsky M, Verner J, Cerna J, et al. Novel CHK1 inhibitor MU380 exhibits significant single-agent activity in TP53-mutated chronic lymphocytic leukemia cells. Haematologica. 2019;104:2443–55.
Johnston JB, Kabore AF, Strutinsky J, Hu X, Paul JT, Kropp DM, et al. Role of the TRAIL/APO2-L death receptors in chlorambucil- and fludarabine-induced apoptosis in chronic lymphocytic leukemia. Oncogene. 2003;22:8356–69.
Rosenwald A, Chuang EY, Davis RE, Wiestner A, Alizadeh AA, Arthur DC, et al. Fludarabine treatment of patients with chronic lymphocytic leukemia induces a p53-dependent gene expression response. Blood. 2004;104:1428–34.
Xue W, Kitzing T, Roessler S, Zuber J, Krasnitz A, Schultz N, et al. A cluster of cooperating tumor-suppressor gene candidates in chromosomal deletions. Proc Natl Acad Sci USA. 2012;109:8212–7.
Chapiro E, Lesty C, Gabillaud C, Durot E, Bouzy S, Armand M, et al. “Double-hit\“ chronic lymphocytic leukemia: an aggressive subgroup with 17p deletion and 8q24 gain. Am J Hematol. 2018;93:375–82.
Bonfiglio S, Sutton LA, Ljungstrom V, Capasso A, Pandzic T, Westrom S, et al. BTK and PLCG2 remain unmutated in one-third of patients with CLL relapsing on ibrutinib. Blood Adv. 2023;7:2794–806.
Secchiero P, Tiribelli M, Barbarotto E, Celeghini C, Michelutti A, Masolini P, et al. Aberrant expression of TRAIL in B chronic lymphocytic leukemia (B-CLL) cells. J Cell Physiol. 2005;205:246–52.
MacFarlane M. TRAIL-induced signalling and apoptosis. Toxicol Lett. 2003;139:89–97.
Kelley RF, Totpal K, Lindstrom SH, Mathieu M, Billeci K, Deforge L, et al. Receptor-selective mutants of apoptosis-inducing ligand 2/tumor necrosis factor-related apoptosis-inducing ligand reveal a greater contribution of death receptor (DR) 5 than DR4 to apoptosis signaling. J Biol Chem. 2005;280:2205–12.
MacFarlane M, Inoue S, Kohlhaas SL, Majid A, Harper N, Kennedy DB, et al. Chronic lymphocytic leukemic cells exhibit apoptotic signaling via TRAIL-R1. Cell Death Differ. 2005;12:773–82.
Irmler M, Thome M, Hahne M, Schneider P, Hofmann K, Steiner V, et al. Inhibition of death receptor signals by cellular FLIP. Nature. 1997;388:190–5.
Olsson A, Diaz T, Aguilar-Santelises M, Osterborg A, Celsing F, Jondal M, et al. Sensitization to TRAIL-induced apoptosis and modulation of FLICE-inhibitory protein in B chronic lymphocytic leukemia by actinomycin D. Leukemia. 2001;15:1868–77.
Kaufmann SH, Steensma DP. On the TRAIL of a new therapy for leukemia. Leukemia. 2005;19:2195–202.
Droin N, Guery L, Benikhlef N, Solary E. Targeting apoptosis proteins in hematological malignancies. Cancer Lett. 2013;332:325–34.
Herbeuval JP, Nilsson J, Boasso A, Hardy AW, Vaccari M, Cecchinato V, et al. HAART reduces death ligand but not death receptors in lymphoid tissue of HIV-infected patients and simian immunodeficiency virus-infected macaques. Aids. 2009;23:35–40.
Acknowledgements
The present research was funded by Roche Diagnostics, Force Hemato (grant reference: 03–2022), French Innovative Leukemia Organization (FILO) group, ACLF (grant 2022) and SIRIC-CURAMUS (Cancer United research Associating Medecine, University and Society; grant reference: INCa-DGOS-INSERM_12560 and INCa-DGOS-INSERM-ITMO Cancer_18010). L.J. and L.D. received PhD fellowships from Fondation ARC and SIRIC-CURAMUS, respectively. L.K.L. received postdoctoral fellowships from Fondation ARC and Fondation de France. L.S. was funded by the FILO group and the SIRIC-CURAMUS. We thank M Yon, E Pramil and C Gabillaud for preliminary assessments. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
FN-K. and SAS designed the study. LJ and EC performed experiments and analyzed data. LD, CD, LS, BG, FD, LKL, DG, CB performed experiments. OT and DR-W provided samples and clinical data. MB performed statistical analyses. FN-K, SAS, EC and LJ wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Jondreville, L., Dehgane, L., Doualle, C. et al. del(8p) and TNFRSF10B loss are associated with a poor prognosis and resistance to fludarabine in chronic lymphocytic leukemia. Leukemia 37, 2221–2230 (2023). https://doi.org/10.1038/s41375-023-02035-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41375-023-02035-3